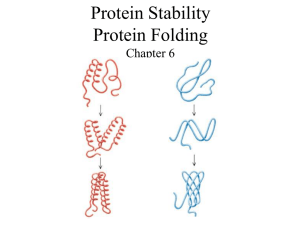
Fold
advertisement
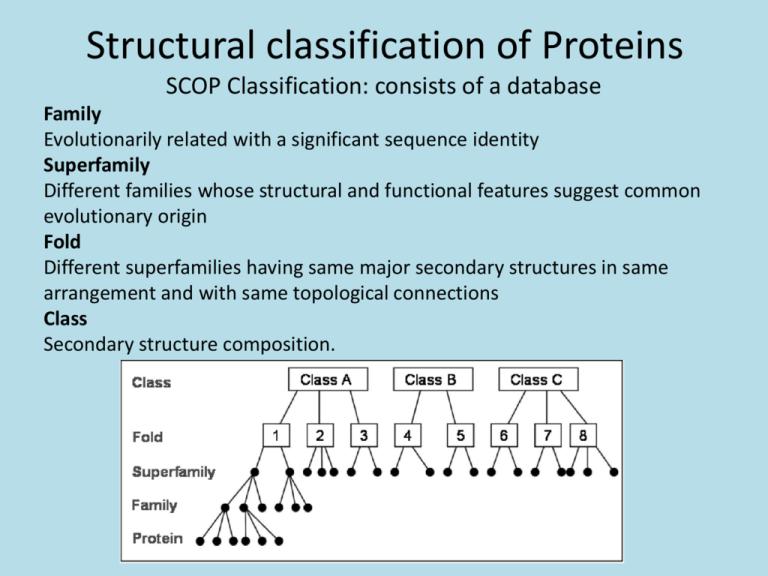
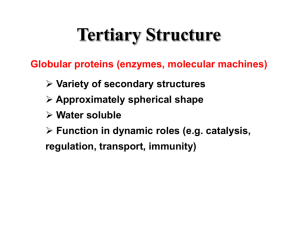
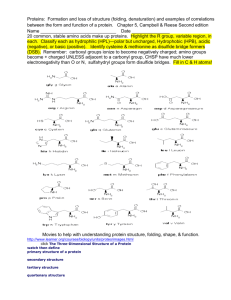
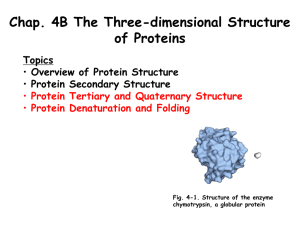
Structural classification of Proteins SCOP Classification: consists of a database Family Evolutionarily related with a significant sequence identity Superfamily Different families whose structural and functional features suggest common evolutionary origin Fold Different superfamilies having same major secondary structures in same arrangement and with same topological connections Class Secondary structure composition. Databases of Structural Classification SCOP: Structural Classification of Proteins CATH: Class, Architecture, Topology and Homologous superfamily FSSP: Families of Structurally Similar Proteins The Protein Folding Problem Given a particular sequence of amino acid residues (primary structure), what will the tertiary/quaternary structure of the resulting protein be? Input: AAVIKYGCAL… Output: 11, 22… => backbone conformation (no side chains yet) But what about the tertiary structure? Folding intermediates • Levinthal’s paradox – Consider a 100 residue protein. If each residue can take only 3 positions, there are 3100 = 5 1047 possible conformations. • Finding a native folded state among all possible configurations can take an enormously long time • Folding must proceed by progressive stabilization of intermediates for fast folding – Molten globules – most secondary structure formed, but much less compact than “native” conformation. – It is an intermediate between the native state and denatured state. Forces driving protein folding It is believed that hydrophobic collapse is a key driving force for protein folding fast reaction; produces molten globule state Hydrophobic core Polar surface interacting with solvent Minimum volume (no cavities) Disulfide bond formation stabilizes Hydrogen bonds Polar and electrostatic interactions Forces driving protein folding Proteins are, in fact, only marginally stable Native state is typically only 5 to 10 kcal/mole more stable than the unfolded form Many proteins help in folding Protein disulfide isomerase – catalyzes shuffling of disulfide bonds Chaperones – break up aggregates and (in theory) unfold misfolded proteins Recall: secondary and tertiary structure of proteins, and quaternary structure Determining Protein structure Coordinates are determined by X-ray crystallography The interaction of x-rays with electrons arranged in a crystal can produce electron-density map, which can be interpreted to an atomic model. Crystal is very hard to grow. Nuclear magnetic resonance (NMR) •Some atomic nuclei have a magnetic spin. •Probe the molecule by radio frequency and get the distances between atoms. •Only applicable to small molecules. PDB: Protein Data Bank Three-dimensional structures of large biological molecules, including proteins and nucleic acids. Contain sequence details, atomic coordinates, crystallization conditions Ab initio Structure Prediction Deriving structures, approximate or otherwise, from sequence. An free energy function to describe the protein •bond energy •bond angle energy •dihedral angle energy •van der Waals energy •electrostatic energy Minimize the function and obtain the structure •An algorithm capable of finding the global minimum of the energy function is be used Not practical in general •Computationally too expensive •Accuracy is poor Ab initio Structure Prediction contd. • It should be kept in mind that native structures exist in a certain solvent environment • The native conformation need not necessarily correspond to the global minimum of free energy. Template based protein structure prediction or also known as: comparative modeling, homology modeling Used where there is a clear sequence relationship between the target structure and one or more known structures. Template based…(contd) The most reliable technique for predicting protein structure Comparing the sequence of the new protein with the sequences of proteins of known structure Strong similarity No strong similarities comparative modeling cannot be used. Components •Protein Structure library (Template library) From PDB, FSSP, SCOP •Scoring Function Sequence compatibility Structure compatibility •Alignment Algorithm •Fold Recognition •Confidence Assessment 3D Model Building Programs: MODELLER, SWISS-MOD, SCWRL, etc Similar performances (from CASP) Best Template? Closest biological function? Environmental factors (pH, ligand, etc) Resolution Choosing family of proteins Using multiple templates Alignment accuracy is everything! Cannot recover from an incorrect alignment Gap placement Try many plausible alignments, and build multiple models Build as many models as possible, and determine the best Consensus Using protein structure analysis program Prediction of protein structure includes: • Protein secondary structure prediction • Protein Phi-Psi angle prediction • Predicting disulphide Bridges • Predicting beta-turns • Domain recognition • Domain boundary detection • Protein structural classification • Mining structural motifs