decisions cellulaires - Département de biologie
advertisement

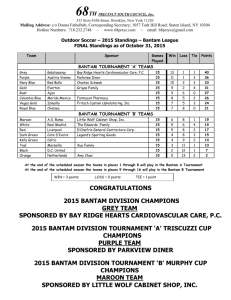
M1 BIOLOGIE DES EUCARYOTES Première Session 2007 DECISIONS CELLULAIRES Durée de l’épreuve: 2h Il s’agit d’un petit travail de synthèse INTRODUCTION The tight control of cell proliferation and cell death is essential to normal tissue development, and the loss of this control is a hallmark of cancers. Coordination between cell proliferation and cell death is essential to maintain homeostasis in multicellular organisms. In Drosophila, these two processes are coordinately regulated during development by the Hippo signaling pathway. The Hippo tumor-suppressor pathway has emerged as a key signaling pathway that controls tissue size in Drosophila. Hippo signaling restricts tissue size by promoting apoptosis and cellcycle arrest, and animals carrying clones of cells mutant for hippo develop severely overgrown adult structures. This pathway is so far composed of two membrane-associated Band 4.1 proteins, Expanded (Ex) and Merlin (Mer), two serine/threonine kinases, Hippo (Hpo) and Warts (Wts), the scaffold proteins Salvador (Sav) which binds to Hpo and Mats, a regulator of Wts activity. Ex, Mer, Hpo, Sav, Wts, and Mats are negative regulators of growth, and loss-of-function mutations in these genes result in dramatically overgrown tissues containing an excess number of cells. It has been proposed that Ex and Mer act upstream of Hpo, which in turn phosphorylates and activates Wts. Wts negatively regulates the transcription of cell-cycle and cell-death regulators such as cycE and diap1 by a mechanism described in the Article 1. All of the components of the Hippo pathway are conserved in humans and several have been implicated in human cancer, in particular Merlin, which in humans is encoded by the Neurofibromatosis type-2 tumor suppressor gene. ARTICLE 1 Huang J et al. Cell. 2005;122(3):421-34. Identification of Yorkie (Yki) as a Wts Binding Protein in a two-hybrid screen for Wts binding proteins. -Yki is most closely related to the human yes-associated protein (YAP). YAP is a potential oncogene in mammals. YAP has been implicated as a transcriptional coactivator, a class of transcriptional regulators that do not bind to DNA themselves but associate with DNA binding transcription factors and supply or stimulate transcriptional activation of the cognate transcription factors. -Activation of Yki leads to massive tissue overgrowth that resembles the loss-of-function phenotype of hpo, sav, or wts whereas cells mutant for Yki grow poorly. -Activated Yki drives growth by stimulating cell proliferation and inhibiting apoptosis. -When Hpo signaling is reduced, for example by mutations in hpo, Yki is hypophosphorylated and active, driving the expression of target genes that promote cell proliferation and suppress apoptosis, resulting in tissue overgrowth. -Yorkie is known to activate Cyclin E (CycE) and the apoptosis inhibitor DIAP1. 1 However, overexpression of these two targets is not sufficient to cause tissue overgrowth. How Yorkie regulates growth, and thus the identities of downstream target genes that mediate these effects of Hippo signaling, are largely unknown. ARTICLES 2 et 3 Nolo et al. Curr Biol. 2006;16(19):1895-904. B J. Thompson and S M. Cohen, Cell 2006; 126(4): 767–774, The bantam locus of Drosophila was identified in a gain-of-function screen for genes that affect tissue growth (Hipfner et al., 2002). bantam encodes a microRNA (miRNA) Experience 1 GMR-Gal4: UAS-hid GMR-Gal4: UAS-hid,UAS-bantam GMR-Gal4: UAS-hid;bantam-/+ SEM images of eyes of adult flies. The genotypes of the animals are indicated above the panels. GMR drives overexpression of UAS transgenes in the eye. Experience 2 Quantification of cell numbers in pupal retinae. Wild-type retinae or retinae with bantam mutant clones had 1.67 cells in such regions, whereas retinae overexpressing Yki had 5.9 +/- 1.0 interommatidial cells and regions that overexpressed Yki but were mutant for bantam had 3.1+/0.5 cells. Error bars represent standard deviations. 2 Experience 3 B A wt GMR:Yki wt GMR:Yki (A) Northern blot probed with radiolabeled anti-bantam oligonucleotide. The same blot is probed with an anti-tRNA radiolabeled oligonucleotide as a loading control. (B) Quantitative-PCR analysis of bantam microRNA levels in wild-type retinae or retinae overexpressing Yki. Experience 4 Wt GMR-Gal4: UAS-bantam SEM images of eyes of adult flies and third-instar eye discs stained with antibody against activated Caspase 3 (Casp). The genotypes of the animals are indicated above the panels. GMR-Gal4 drives overexpression of UAS transgenes in the eye. 3 Experience 5 (A–C) show SEM images of eyes of adult flies and (A’– C’) show higher-magnification images of the panels above them. (A’’_ C’’) show third-instar eye discs stained with antibody against Drice to mark apoptotic cells. The genotypes of the animals are indicated above the panels. (A-A’’: GMR-Gal4, B-B’’:GMR-Gal4:UAS- hpo, C-C’’:GMR-Gal4:UAS- hpo,UAS- ban - Vous rappellerez le principe des expériences permettant la sur-expression ou l’expression ectopique d’un gène chez la drosophile. - Vous donnerez une conclusion de l’ensemble des résultats présentés ci-dessus en insistant sur le rôle de bantam. - Vous proposerez un modèle, accompagné d’une légende, de la voie de contrôle de la taille des tissus par Hippo en utilisant les signes conventionnels : signifie active et signifie réprime Un rappel des principaux acteurs moléculaires impliqués est donné sur la page 5 4 APOPTOSIS CELL GROWTH PROLIFERATION Bantam ? 5