Supplementary Material (doc 770K)
advertisement
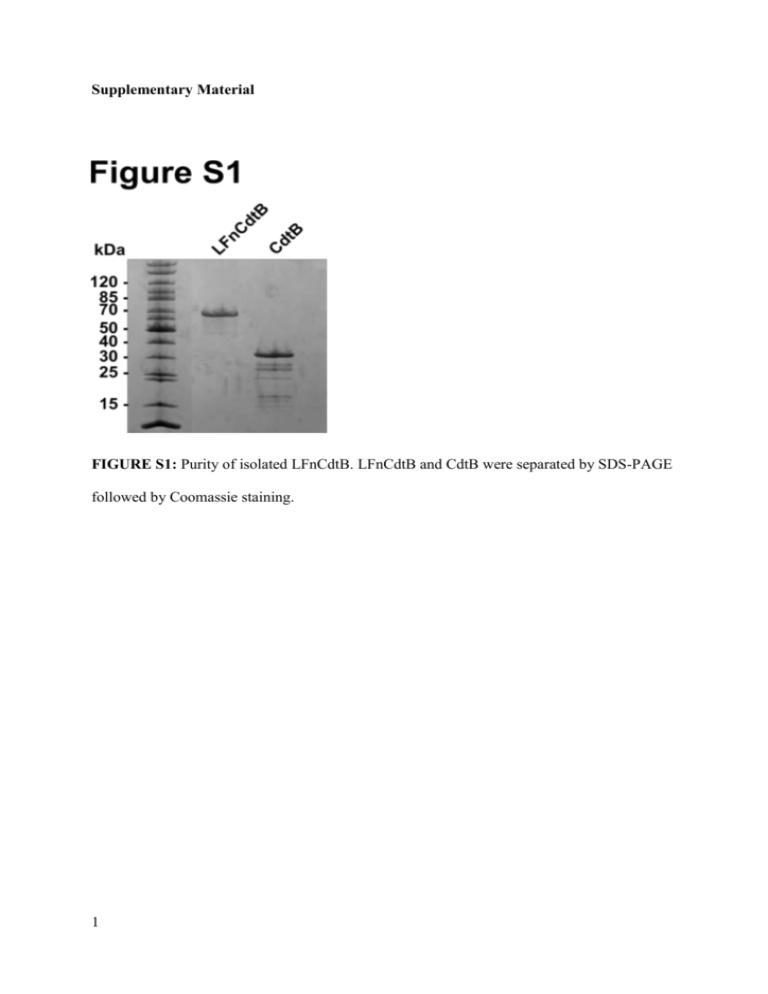
Supplementary Material FIGURE S1: Purity of isolated LFnCdtB. LFnCdtB and CdtB were separated by SDS-PAGE followed by Coomassie staining. 1 FIGURE S2: Electrospray ionization mass spectrometry of purified LFnCdtB. The mass determined by deconvolution of the mass/charge plot was 59,712, which compares closely to the calculated mass of 59,709. 2 FIGURE S3: In-vitro DNase activity assay for LFnCdtB. 25 pmol CdtB, LFnCdtB or 1 U DNase I were incubated with 1 µg purified plasmid pET19b and incubated for 2 h. Separation of DNA was performed by agarose gel electrophoresis. Additional linearized DNA band indicates CdtB enzymatic activity. 3 FIGURE S4: Analysis of blood enzyme levels after 6 injections (days 5, 7, 9, 12, 14, 16) of 100 µg PA-L1 + 100 µg LFnCdtB or PBS into the peritoneum of C57BL/6 mice. Each group comprised of 5 mice and for statistical analysis, data of 15 further mice treated similarly with PBS were applied. Statistical significance was calculated by an unpaired t-test and significant p values are presented. 4 Supplementary Materials and Methods Cloning of LFnCdtB – The cDNA CdtB from H. ducreyi was amplified by PCR to generate the suitable restriction sites MluI and XmaI for cloning CdtB into the expression plasmid FP59AGGpYS for B. anthracis expression. The following sequence was generated: Signal sequence (from B. anthracis protective antigen): MKKRKVLIPLMALSTILVSSTGNLEVIQ LFn (GenBank Accession Number AAA79216.1): AGGHGDVGMHVKEKEKNKDENKRKDEERNKTQEEHLKEIMKHIVKIEVKGEEAVK KEAAEKLLEKVPSDVLEMYKAIGGKIYIVDGDITKHISLEALSEDKKKIKDIYGKDAL LHEHYVYAKEGYEPVLVIQSSEDYVENTEKALNVYYEIGKILSRDILSKINQPYQKFL DVLNTIKNASDSDGQDLLFTNQLKEHPTDFSVEFLEQNSNEVQEVFAKAFAYYIEPQH RDVLQLYAPEAFNYMDKFNEQEINLS Peptide linker: TRSGENLYFQS CdtB (PubMed Data Base Accession Number 1SR4_B) : NLSDFKVATWNLQGSSAVNESKWNINVRQLLSGEQGADILMVQEAGSLPSSAVRTS RVIQHGGTPIEEYTWNLGTRSRPNMVYIYYSRLDVGANRVNLAIVSRRQADEAFIVH SDSSVLQSRPAVGIRIGTDVFFTVHALATGGSDAVSLIRNIFTTFNSSSSPPERRVYSW MVVGDFNRAPANLEVALRQEPAVSENTIIIAPTEPTHRSGNILDYAILHDAHLPRREQ ARERIGASLMLNQLRSQITSDHFPVSFVRDR Expression and Purification of LFnCdtB – LFnCdtB was basically expressed and purified as described earlier 1. Expression was done in plasmid-cured and non-infectious, protease-deficient B. anthracis strain BH460 2. Following after anion exchange chromatography, cation exchange chromatography was used as the final purification step. After elution from Phenyl Sepharose samples were pooled, dialyzed against 20 mM Tris, pH 5 8, 0.5 mM EDTA overnight and loaded on a Q-Sepharose Fast Flow column for purification with an Äkta chromatography system (GE Healthcare, Waukesha, WI) and eluted using a linear gradient of 20 mM Tris, pH 8, 0.5 mM EDTA, 0.5 M NaCl. Fractions containing LFnCdtB were dialyzed against 20 mM citric acid, pH 6.8, 0.5 mM EDTA overnight and loaded on an S-Sepharose Fast Flow column for purification with an Äkta chromatography system and eluted using a linear gradient of 20 mM citric acid, pH 6.8, 0.5 mM EDTA, 0.5 M NaCl. Fractions containing LFnCdtB were dialyzed against 5 mM HEPES, pH 7.2, 0.5 mM EDTA, concentrated (Amicon Ultrafiltration devices, 30 kDa molecular weight cutoff, Millipore, Billerica, MA), filter sterilized, and stored in aliquots at -80 °C. With a culture volume of 5 L a total yield of 17 mg or 3.4 mg LFnCdtB / mL culture medium was achieved. Purified LFnCdtB was analyzed by electrospray ionization mass spectrometry to confirm that the mass matched the calculated mass from its sequence. DNase Activity Assay – 1 µg of plasmid DNA was incubated with 25 pmol of CdtB or LFnCdtB or 1 µL DNase I at 37 °C for 2 h in a total volume of 15 µL 25 mM HEPES, pH 7.2, 50 mM MgCl2. Samples were analyzed on a 0.7 % agarose gel. Cell Culture – CHO K1 cells were maintained in modified Eagle’s medium alpha with Glutamax-1 (Gibco, Life Technologies, Grand Island, NY). All other cells were maintained in Dulbecco’s modified Eagle’s medium with Glutamax-1 (Gibco, Life Technologies, Grand Island, NY). All media were supplemented with 20 % (LL3 and B16/BL6 cells) or 10 % (all other cells) fetal bovine serum (Gibco, Life Technologies, Grand Island, NY) and 50 µg/mL gentamicin (Quality Biological, Gaithersburg, MD). Cytotoxicity Assays – Dose-response curves for the combination of PA + LFn fusion proteins (LFnCdtB and FP59AGG) or CdtB were obtained by incubation on cells for 72 h. The cells (5000 cells per well in 100 µL medium) were seeded in 96-well plates and incubated at 37 °C overnight before addition of PA and the LFn fusion proteins. PA and LFn 6 fusion proteins were added in 100 µL to achieve final concentrations of 250 ng/mL for PA (both wildtype PA and PA-L1) and 10 fM-100 nM of LFn fusion proteins or CdtB, depending on the tested cell lines. The cells were incubated a further 72 h and cell survival was determined in an 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma Aldrich, St. Louis, MO) assay as described elsewhere (Ub paper). Data Analysis – The 50 % survival indices (SI50, 50 % cell survival in comparison to untreated controls) values for cytotoxicity analyses were obtained from a nonlinear regression curve fit using GraphPad Prism 5.02. The method used for the nonlinear regression curve fit was “log(inhibitor) versus normalized response” (by using a least square fit). Analyses of cell cycle arrest and TUNEL-positive cells were performed by using FlowJo. The percentage of cells in the G0/G1 phase and in the G2/M phase was analyzed by using the cell cycle analysis of FlowJo. For statistical analysis of blood enzyme levels, data of 15 mice treated with PBS for other experiments were included and compared by using the unpaired t-test (GraphPad Prism 5.02). Reference List 1. Park S, Leppla SH. Optimized production and purification of Bacillus anthracis lethal factor. Protein expression and purification 2000 Apr; 18(3): 293-302. 2. Pomerantsev AP, Pomerantseva OM, Moayeri M, Fattah R, Tallant C, Leppla SH. A Bacillus anthracis strain deleted for six proteases serves as an effective host for production of recombinant proteins. Protein expression and purification 2011 Nov; 80(1): 80-90. 7