Restriction Mapping with Branch and Bound Algorithm
advertisement

Report on “Solving Restriction mapping Problem using Branch and
Bounds Algorithm”
Abstract
Restriction enzymes which are developed by Bacteria are used to cut specific sites of DNA called
‘restriction sites’. Every bacteria has it’s own specific recognition and cut sites. These sites are
usually 4-6 base pair long. So A DNA molecule with n number of restriction sites (complete digest
set) produce n+1 number of fragments. If the
sample of DNA is exposed to the restriction
enzyme for only a limited amount of time to prevent it from being cut at all restriction sites. So we
generate the set of all possible restriction fragments between every two cuts. This set of
fragments are called ‘partial digest set’ is used to determine the positions of the restriction sites in
the DNA sequence. Before the DNA sequencing reaction process were invented, calculating the
complete digest set from partial digest set was the only process to know the restriction sites in
DNA and so as the sequence of base pairs. So an experiment is done based on an algorithm
called Branch and bound algorithm. With a given set of partial digest, the program gives the
complete digest set. The only exception for this program is for a same partial digest set there are
two different complete digest set called Homometric set. This program involves Recursive
Selection Sort having the running time O(n2) in average case.
Introduction
Restriction Mapping is the process of getting structural information on a sequence of
DNA by the help of Restriction enzymes. Restriction enzymes are endonuclease that
recognize specific base in DNA called ‘Restriction sites’ and make cleavages. As DNA
sequencing technology were discovered recently, few years ago restriction maps
became powerful research tools in molecular biology by helping the location of genetic
markers. The distance between two restriction sites can be calculated experimentally by
gel electrophoresis. So a DNA molecule can be either fully digested by restriction
enzymes, or partially digested which cut the DNA with a probability less than 1, there
by producing all the possible fragments of two consecutive sites. The Objective of this
project is to obtain the set of complete digest from a given partial digest sets.
Restriction digest problem can be solved by implementing various algorithms
1. Brute Force Algorithm- This is exhaustive search and efficient but very slow and
need memory in each step.
2. Another Brute Force Algorithm- This is more efficient, faster than Brute Force but
still slow.
3. Branch and Bound Algorithm- Most efficient among three and relatively faster
than the two above.
Branch and Bound Algorithm
PartialDigest(L):
width <- Maximum element in L
DELETE(width, L)
X <- {0, width}
PLACE(L, X)
1. PLACE(L, X)
2.
if L is empty
3.
output X
4.
return
5. y <- maximum element in L
6. Delete(y,L)
7. if D(y, X ) Í L
8.
Add y to X and remove lengths D(y, X) from L
9.
PLACE(L,X )
10.
Remove y from X and add lengths D(y, X) to L
11. if D(width-y, X ) Í L
12.
Add width-y to X and remove lengths D(width-y, X) from L
13.
PLACE(L,X )
14.
Remove width-y from X and add lengths D(width-y, X ) to L
15. return
Implementation of above algorithm is done by using Java. The following are the
methods involved in solving the problems. It has one main method with ten different
class methods to get the solution. The data application process flow is detailed in fig 1.
main :This is the main method which first calls assighnData method that
validates and accept the user’s input as partial digest set. Then it include 0
and last element in the Complete digest set and remove the same from
partial digest set by deletePD method. Then it calls placeL and print the
array.
placeL : this is the major method which calls itself recursively and calls
other methods. First it takes one element at a time from right side in the
partial digest array. Build the distance array matrix for that number by
‘distanceArray’. Then call ’ifDExistInL’ method to check if all the distance
array are exist in partial digest set. If that is true, add the number in
complete digest set and delete the same from partial digest set. If that is
not true, then subtract the no from the highest no of complete digest set.
Again build the distance matrix for this new no and check whether all are
present in PD set. If yes then include the new no in CD. Else move to next
number. This method print every iteration and made the process more
clearer.
ifDExistInL: This is a boolean method compare each and every element
of PD with the distance matrix and return either true or false accordingly.
assignData: This is another important method which first validate the total
number of PD given by user. It can be achieved by following formula
numOfCD = (int)(Math.sqrt(numOfPD * 2.0)+1)
As it is known that numOfPD = [numOfCD(numOfCD-1)] /2, so the
method again rechecking whether the calculated CD is a valid one or not.
Then it takes the PD number separated by comma and put it in an array
by using string functions.
distanceArray: This is just calculate the distance matrix for any number
passed as parameters.
deletePD: This method called by placeL method. It delete every elements
of distance matrix if they are present in PD set for validating a number. But
it delete a number only once if the number has repetition, as the repeated
number may be generated by some other number.
addCD: if the number validation is true in placeL method, then that
number is added to the array of complete digest.
sortPD: sorting the element after the user input and after every delete
statement on partial digest set.
sortCD: sorting the elements after every addCD methods called.
printArray: It just print the existing array with curly ({}) brackets.
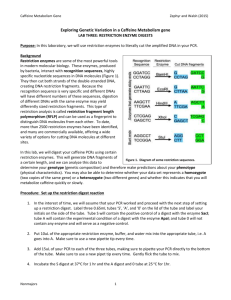
Start
Read comma
separated data, split
to array, calculate #
complete digest
Read Data
Fail
Validate
Data
Pass
Evaluate next
element
No
Delete from PD
& Store in CD
If PD =
NULL
Yes
Yes
If Dist Array
exists in PD
Show
Result
In CD
Compute
Distance
Array
Fig 1: Application Process Flow
Result
The java program gives final result with set of complete digest.
This Branch and bound algorithm work very well in high quality PDP data. Unlike it’s
corresponding PDP algorithm as BruteForce and AnotherBruteForce
algorithm, this
Branch and Bound one is very fast and efficient too. At each point we examine two
alternatives left or right, ruling out the obvious incorrect decision. This algorithm is very
fast for most instance of PDP because only one of the two alternatives either left or
right is viable at any step.
Let T(n) be the maximum time Partial Digest takes to find the solution for an n-point
instance of the PDP. If there is only one viable alternatives at every step, then the
program steadily reduce the size of the problem by one and calls itself recursively, it is
also called RECURSIVE SELECTION SORT.
T(n) = T(n-1) + n,
T(1) = 1,
Therefore,
T(n) = n + T(n-1)
= n + (n-1) + T(n-2)
= n + (n-1) + (n-2) +…….+3+2+T(1)
= O(n2)
So the upper bound of the algorithm is quadratic time that is : O(n2) ,
However if there are two alternatives, then
T(n) = 2T(n-1) + n
2n-1
So, the Big O for this is an exponential algorithm.
Exception
One of the exception of this program is with same set of partial digest set, there are two
possible complete digest set. These two sets are called Homometric set. There is no
solution for this kind of problem.
Future Works
The partial digest algorithm can be applied in following two problems.
Motif finding problem: Given a set of DNA sequences, find a motif , one from
each sequence, that maximizes the consensus score.
Median String problem: Given a set of DNA sequences, find a median string.
Both the above problem can be solved by Branch and Bound algorithm. These
application shows more efficiency over the exhaustive BruteForce one in speed, but
unfortunately it always goes exponential time to find.
Conclusion
As it is understood that the partial digest problem can be solved by various algorithm,
like BruteForce, AnotherBruteForce and Branch and Bound algorithm. The later one is
significantly faster than previous two algorithm with very good PDP data set. Although
this problem shows an exception when the data set is homometric that is two possible
complete digest set from same partial digest set. The running time is usually O(n2) in
best or average case but it becomes exponential in worst case.
Reference
Neil C. Jones and Pavel A. Pevzner – An Introduction to Bioinformatics
Algorithm.
K.N. King- An introduction to Java Programming.