Lab 5, Conceptor 4 answer key - Cal State LA
advertisement

Biol/Chem 444
Drs. Krug & McCurdy
Spring, 2008
Molecular Conceptor Exercise 4
One of the most successful applications of receptor-based drug design has been the
development of compounds that selectively inhibit the HIV virus. An estimated 40 million
people worldwide are HIV+; in parts of Africa, more than 25% of the adult population carries
the virus. HIV is a global pandemic that will soon surpass malaria as the leading source of
mortality from infectious disease. In the 1980’s, the only drugs available to treat HIV were
nucleoside analogues such as AZT that blocked the reverse transcription of viral RNA.
These drugs suffered from the rapid appearance of resistant viral enzymes.
Beginning in the mid-90’s, efforts were made to target a different viral enzyme, the HIV
protease. All the different HIV proteins are initially translated as one long protein, which
is then cut by the protease enzyme into a bunch of separate proteins (analogy: A Hershey’s
bar comes as many squares attached together, which you can break off and eat individually.)
Without the protease, HIV cannot complete its lifecycle, even after successfully infecting a
cell. From the crystal structure of the HIV protease active site, rational drug design was used
to invent compounds that would block the protease by optimally filling all the pockets and
making all the H-bonds a natural substrate would make within the catalytic site.
You will have 1 hour 15 minutes to complete this activity; we will then work on the primary
literature assignment. If you finish early, you may want to review the paper for today.
[D2.9.1] HIV protease background. Review the background and make sure you
understand what the HIV protease enzyme does. Proteases cleave only a peptide bond that
is properly positioned in their active site, breaking the backbone of substrate proteins. The
chemical features of the peptide bond are essential for a protease to catalyze this reaction;
make sure you know what a peptide bond is.
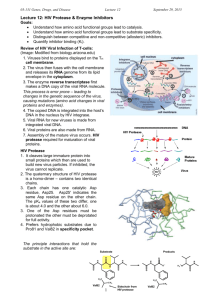
[D2.9.3] Examine the crystal structure of a peptide substrate bound to the catalytic site of the
protease. Define dimer and -carbon.
dimer: protein complex that functions when two identical subunits
(monomers) bind together
a-carbon: the backbone carbon of a peptide that is not involved in
the peptide bond, to which the side chain is attached
1. [D2.9.6] Inhibitor structure. Why can’t the inhibitor pictured here be cleaved by the
protease enzyme, like its normal substrate would be? What is different about the inhibitor’s
structure? Sketch a peptide bond such as HIV protease would cleave in its natural protein
substrate; below it sketch the corresponding part of the inhibitor that protease cannot cut.
Inhibitor is missing the carbonyl, thus does not have a peptide bond in
the central position; without this bond, the inhibtor cannot serve as a
substrate for the HIV protease enzyme (it cannot be hydrolyzed, i.e. cut)
1
Biol/Chem 444
Spring, 2008
Drs. Krug & McCurdy
2. [D2.9.8, button 2] Through specific binding interactions in the active site, HIV protease
inhibitors are able to hold the protease enzyme locked into its “closed” conformation. This
section will enable you to examine the intermolecular forces involved.
a. Which functional groups on the inhibitor are involved? For each group, state whether it
is an H-bond donor or acceptor, and what molecule is donating or receiving its H-bond.
Each carbonyl of the central pair of carbonyls acts as an H-bond
acceptor for a proton from the bound water molecule.
b. Which residues on the protease are involved? State whether they are H-bond donors or
acceptors, and what molecule or functional group is donating or receiving their H-bonds.
Each of the 2 Isoleucines (Ile 50, Ile 50’) are also involved. Each
acts as an H-bond donor from its backbone amide to the oxygen of the
bound water.
c. [screen 3] For each residue, state whether the backbone, side chain, or both participate
in H-bonds with the inhibitor. {No need to repeat the same residue on each monomer.}
Asp 29 – backbone NH acts as H-bond donor
sidechain carboxylate acts as acceptor
Gly 27 – backbone carbonyl acts as acceptor
Gly 48 – backbone carbonyl acts as acceptor
3. [D2.9.9 to D2.9.10] Using information on the protease active site, improvements were
made to the initial lead compound, resulting in the drug A-77003.
a. What makes this molecule a better inhibitor than previous drugs? (There are 2
improvements that were tailor-made to accommodate features of the protease active site).
(1) The diol moiety (pair of adjacent hydroxyls) provides a pair of
H-bonds, with each –OH acting as an H-bond donor to the side chain
carboxylate of Asp 25 + 25’
(2) Bulky hydrophobic groups (aromatic rings, isopropyl groups) fill
the 4 hydrophobic pockets
b. The drug A-77003 is called “peptidic” or peptide-like, but it is not a “real” peptide. In
what way is this molecule different from an actual peptide? Why is this important?
The central bond is not a peptide bond, but rather a diol; this is
not a substrate for the protease, so it cannot be cleaved. Instead,
the inhibitor will be block the active site and not be broken down.
There are peptide binds on either side of the diol, but these will
not be positioned in the catalytic site, so they also will not be
cleaved by the enzyme.
2
Biol/Chem 444
Spring, 2008
Drs. Krug & McCurdy
4. [D2.9.11] a. What pharmacophore was proposed based on the A77003/HIV protease
complex? What structure turned up as a hit in the database search using this pharmacophore?
b. What functional groups on the hit are essential for activity? What part of the normal
substrate do they mimic or replace?
4. [D2.9.13, buttons 1-5] One of the principles that emerged from receptor-based analysis
of HIV protease is that drugs will bind better if they can replace the water molecule that is
bound to Ile50 + 50’, with a functional group that can maintain the H-bonding interactions
that the water molecule participated in. (In other words, incorporate into the structure a
group that will displace the water but still make the required H-bonds.) What groups were
put on the ring to replace the water? Why is it advantageous to replace the water?
One molecule has a methoxy group, whereas the other molecule
has a carbonyl in the same place; both function as H-bond
acceptors, replacing the water.
5. [D2.9.14, buttons 1-4] These screens show how a 6-membered ring (yellow) was used as
a base for attaching various functional groups, to try to optimally fill all the available binding
regions of the active site. Switching to a 7-membered ring (blue) solved 3 problems that
were encountered with the 6-membered-ring compounds. Working through screen
[D2.9.15], explain what these 3 improvements were.
(1) better positioning of the diol for optimal H-bond donation to
Asp 25/25’
(2) proper stereochemistry to position 4 phenyl groups into the 4
hydrophobic pockets
(3) position of a carbonyl moiety to replace bound water from
previous structures; the carbonyl accepts H bonds from the amide of
Ile 50 + 50’, instead of the displaced water.
3