FOXC1 mutations are involved in autosomal dominant and
advertisement

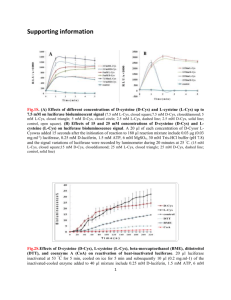
1 SUPPLEMENTARY FIGURE LEGENDS 2 3 Supplementary Figure 1. The rs6533526:C>T, c.*454C>T PITX2 variant decreases 4 luciferase activity. A) The c.*454C>T variant alters the predicted target sequence for 5 hsa-miR-548p. The arrow indicates the position of the mutant nucleotide. The target 6 miRNA was predicted using the MicroSNiPer software. The vertical lines and dots in 7 the predicted miRNA target sequence indicate perfect Watson-Crick and G:U wobble 8 pairings, respectively. Note that the mutant nucleotide forms a perfect Watson-Crik T-A 9 pairing with the has-miR-548p seed sequence. B) Scheme of the cDNA constructs used 10 to assay c.*454C>T associated activity. The 3’-end of the firefly luciferase coding 11 region (1652 nucleotides), cloned into the pMirTarget vector, was coupled with either 12 the wild type or mutant PITX2 3’-UTR (nucleotides c.*1 to c.*737). The numbers below 13 the cartoon indicate nucleotide positions. C) The influence of the c.*454C>T variant on 14 luciferase activity. HEK-293T cells were transfected with either the wild-type or 15 c.*454C>T constructs fused to firefly luciferase. After a 48-hour incubation, luciferase 16 activity was measured as described in the Materials and Methods section. RFP, encoded 17 by the pMirTarget vector, and endogenous LDH were detected by Western blot as 18 transfection and loading controls, respectively. The error bars correspond to the SD of 19 three independent experiments carried out in triplicate. The asterisks indicate 20 statistically significant differences with respect to the control. p values were obtained 21 using a one-way ANOVA followed by Tukey’s multiple-comparison test: p<0.05 (*); 22 p<0.01 (**); p<0.001 (***). 23 24 Supplementary Figure 2. The rs79691946:C>T, p.(P297S) and p.(G380Rfs*144) 25 variants do not alter FOXC1 DNA-binding. (A) The effect of FOXC1 variants on 26 DNA-binding specificity was assessed by EMSA. The oligonucleotides corresponding 1 1 to the FOXC1-binding sequence labeled with biotin at the 5'-end were incubated with 2 10 µg of nuclear extracts from HEK-293T cells transfected with the indicated variants 3 of recombinant FOXC1. The extracts were separated on a 10% nondenaturing 4 polyacrylamide gel electrophoresis and were transferred to a positively charged nylon 5 membrane (Hybond-N+, Amersham). FOXC1-oligonucleotide complexes were 6 visualized by incubation with streptavidin-HRP conjugate and chemiluminescent 7 detection (Chemiluminescent EMSA kit, Thermo Scientific). Unlabeled competitor 8 oligonucleotides were pre-incubated at increasing concentrations from 0.2 to 10 pmol (1 9 to 50-fold excess) with the labeled probe. The arrowhead indicates the position of full- 10 length FOXC1-DNA complexes. (B) The percentage of band volume corresponding to 11 protein-oligonucleotide complexes was estimated by densitometry analysis as indicated 12 in the Materials and Methods section. The error bars correspond to the SD of three 13 independent experiments carried out in triplicate. NT: non-transfected cells (negative 14 control). The asterisks indicate statistically significant differences with respect to the 15 control. p values were obtained using a one-way ANOVA followed by Tukey’s 16 multiple-comparison test: p<0.001 (***). 17 18 Supplementary Figure 3. Subcellular localization of FOXC1 variants associated 19 with congenital glaucoma. The cDNA constructs encoding the indicated recombinant 20 versions of FOXC1 were transiently expressed in HEK-293T cells. The recombinant 21 proteins were detected by fluorescent immunocytochemistry with an anti-myc antibody 22 (Santa Cruz Biotechnology). Nuclei were visualized using fluorescent DAPI staining. 23 Original magnification: X600. 24 2 1 2 3 SUPPLEMENTARY REFERENCES 4 1. Saleem RA, Banerjee-Basu S, Berry FB, Baxevanis AD, Walter MA: Analyses of the 5 effects that disease-causing missense mutations have on the structure and function of 6 the winged-helix protein FOXC1. AmJHumGenet 2001; 68: 627-641. 7 2. Sanchez-Sanchez F, Martinez-Redondo F, Aroca-Aguilar JD, Coca-Prados M, 8 Escribano J: Characterization of the intracellular proteolytic cleavage of myocilin 9 and identification of calpain II as a myocilin-processing protease. JBiolChem 2007; 10 11 12 282: 27810-27824. 3. Sigrist CJ, de Castro E, Cerutti L et al: New and continuing developments at PROSITE. Nucleic Acids Res 2013; 41: D344-347. 13 4. Barenboim M, Zoltick BJ, Guo Y, Weinberger DR: MicroSNiPer: a web tool for 14 prediction of SNP effects on putative microRNA targets. HumMutat 2010; 31: 1223- 15 1232. 16 5. Thompson JD, Higgins DG, Gibson TJ: CLUSTAL W: improving the sensitivity of 17 progressive multiple sequence alignment through sequence weighting, position- 18 specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22: 4673- 19 4680. 20 6. Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB: Protein disorder 21 prediction: implications for structural proteomics. Structure 2003; 11: 1453-1459. 22 7. Linding R, Russell RB, Neduva V, Gibson TJ: GlobPlot: Exploring protein 23 sequences for globularity and disorder. Nucleic Acids Res 2003; 31: 3701-3708. 24 8. Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD: An enhanced 25 computational platform for investigating the roles of regulatory RNA and for 26 identifying functional RNA motifs. BMC Bioinformatics 2013; 14 Suppl 2: S4. 3 1 4