Conserved primer sequences for PCR amplification and sequencing
advertisement
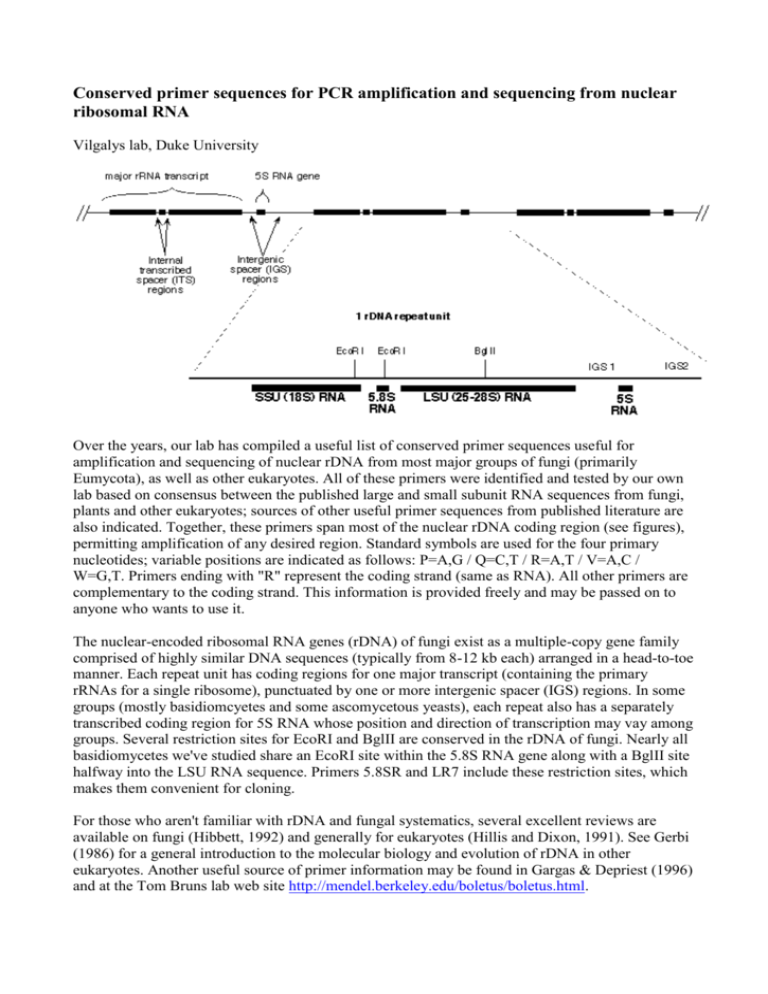
Conserved primer sequences for PCR amplification and sequencing from nuclear ribosomal RNA Vilgalys lab, Duke University Over the years, our lab has compiled a useful list of conserved primer sequences useful for amplification and sequencing of nuclear rDNA from most major groups of fungi (primarily Eumycota), as well as other eukaryotes. All of these primers were identified and tested by our own lab based on consensus between the published large and small subunit RNA sequences from fungi, plants and other eukaryotes; sources of other useful primer sequences from published literature are also indicated. Together, these primers span most of the nuclear rDNA coding region (see figures), permitting amplification of any desired region. Standard symbols are used for the four primary nucleotides; variable positions are indicated as follows: P=A,G / Q=C,T / R=A,T / V=A,C / W=G,T. Primers ending with "R" represent the coding strand (same as RNA). All other primers are complementary to the coding strand. This information is provided freely and may be passed on to anyone who wants to use it. The nuclear-encoded ribosomal RNA genes (rDNA) of fungi exist as a multiple-copy gene family comprised of highly similar DNA sequences (typically from 8-12 kb each) arranged in a head-to-toe manner. Each repeat unit has coding regions for one major transcript (containing the primary rRNAs for a single ribosome), punctuated by one or more intergenic spacer (IGS) regions. In some groups (mostly basidiomcyetes and some ascomycetous yeasts), each repeat also has a separately transcribed coding region for 5S RNA whose position and direction of transcription may vay among groups. Several restriction sites for EcoRI and BglII are conserved in the rDNA of fungi. Nearly all basidiomycetes we've studied share an EcoRI site within the 5.8S RNA gene along with a BglII site halfway into the LSU RNA sequence. Primers 5.8SR and LR7 include these restriction sites, which makes them convenient for cloning. For those who aren't familiar with rDNA and fungal systematics, several excellent reviews are available on fungi (Hibbett, 1992) and generally for eukaryotes (Hillis and Dixon, 1991). See Gerbi (1986) for a general introduction to the molecular biology and evolution of rDNA in other eukaryotes. Another useful source of primer information may be found in Gargas & Depriest (1996) and at the Tom Bruns lab web site http://mendel.berkeley.edu/boletus/boletus.html. Small subunit RNA (17-18S) primer sequences Large subunit RNA (25-28S) and IGS primer sequences Internal transcribed spacer (ITS) region primers Intergenic spacer (IGS) primers (including 5S RNA primer sequences for basidiomycete fungi) back to the top of this page Small subunit RNA (SR) primers: Primer name Sequence (5'-->3') Position within S. cereviseae 17S RNA BMB-'A' GRATTACCGCGGCWGCTG 580-558 BMB-'B' CCGTCAATTCVTTTPAGTTT 1146-1127 BMB-'C' ACGGGCGGTGTGTPC 1638-1624 BMB-BR CTTAAAGGAATTGACGGAA 1130-1148 BMB-CR GTACACACCGCCCGTCG 1624-1640 SR1R TACCTGGTTGATQCTGCCAGT 1-21 SR1 ATTACCGCGGCTGCT 578-564 SR2 CGGCCATGCACCACC 1277-1263 SR3 GAAAGTTGATAGGGCT 318-302 SR4 AAACCAACAAAATAGAA 838-820 SR5 GTGCCCTTCCGTCAATT 1146-1130 SR6 TGTTACGACTTTTACTT 1760-1744 SR6R AAGWAAAAGTCGTAACAAGG 1744-1763 SR7 GTTCAACTACGAGCTTTTTAA 617-637 SR7R AGTTAAAAAGCTCGTAGTTG 637-617 SR8R GAACCAGGACTTTTACCTT 732-749 SR9R QAGAGGTGAAATTCT 896-910 SR10R TTTGACTCAACACGGG 1181-1196 comments NS1 GTAGTCATATGCTTGTCTC NS2 GGCTGCTGGCACCAGACTTGC NS3 GCAAGTCTGGTGCCAGCAGCC NS CTTCCGTCAATTCCTTTAAG (similar to BMB-B) NS5 AACTTAAAGGAATTGACGGAAG (is similar to BMB-BR) NS6 GCATCACAGACCTGTTATTGCCTC NS7 GAGGCAATAACAGGTCTGTGATGC NS8 TCCGCAGGTTCACCTACGGA BMB = "universal" SSU primers developed by Lane et al., 1985 SR = primers developed by Vilgalys lab NS = primers described by White et al., 1990 back to the top of this page Large subunit RNA (25-28S) primer sequences Note: most molecular systematics studies only utilizethe first 600-900 bases from the LSU gene, which includes three divergent domains (D1, D2, D3) that are among the most variable regions within the entire gene (much of the LSU is invariant even across widely divergent taxa). Most of the data in our Agaricales LSU database consists of the first 900 bases from the LSU gene (we typically amplify using primers 5.8SR + LR7, followed by sequencing using primers LR5, LR16, LR0R, and LR3R). Primer name 5.8S Sequence (5'-->3') comments CGCTGCGTTCTTCATCG Position within S. cereviseae rRNA 51-35 (5.8S RNA) 5.8SR TCGATGAAGAACGCAGCG 34-51 (5.8S RNA) contains EcoRI site LR0R ACCCGCTGAACTTAAGC 26-42 LR1 GGTTGGTTTCTTTTCCT 73-57 contains EcoRI site LR2 TTTTCAAAGTTCTTTTC 385-370 LR2R AAGAACTTTGAAAAGAG 374-389 LR3 CCGTGTTTCAAGACGGG 651-635 LR3R GTCTTGAAACACGGACC 638-654 LR4 ACCAGAGTTTCCTCTGG 854-838 LR5 TCCTGAGGGAAACTTCG 964-948 LR6 CGCCAGTTCTGCTTACC 1141-1125 LR7 TACTACCACCAAGATCT 1448-1432 contains BglII site LR7R GCAGATCTTGGTGGTAG 1430-1446 contains BglII site LR8 CACCTTGGAGACCTGCT 1861-1845 LR8R AGCAGGTCTCCAAGGTG 1845-1861 LR9 AGAGCACTGGGCAGAAA 2204-2188 LR10 AGTCAAGCTCAACAGGG 2420-2404 LR10R GACCCTGTTGAGCTTGA 2402-2418 LR11 GCCAGTTATCCCTGTGGTAA 2821-2802 LR12 GACTTAGAGGCGTTCAG 3124-3106 LR12R CTGAACGCCTCTAAGTCAGAA 3106-3126 LR13 CGTAACAACAAGGCTACT 3357-3340 LR14 AGCCAAACTCCCCACCTG 2616-2599 LR15 TAAATTACAACTCGGAC 154-138 LR16 TTCCACCCAAACACTCG 1081-1065 LR17R TAACCTATTCTCAAACTT 1033-1050 LR20R GTGAGACAGGTTAGTTTTACCCT 2959-2982 LR21 ACTTCAAGCGTTTCCCTTT LR22 CCTCACGGTACTTGTTCGCT back to the top of this page 424-393 364-344 Internal transcribed spacer (ITS) region primers The ITS region is now perhaps the most widely sequenced DNA region in fungi. It has typically been most useful for molecular systematics at the species level, and even within species (e.g., to identify geographic races). Because of its higher degree of variation than other genic regions of rDNA (SSU and LSU), variation among individual rDNA repeats can sometimes be observed within both the ITS and IGS regions. In addition to the standard ITS1+ITS4 primers used by most labs, everal taxon-specific primers have been described that allow selective amplification of fungal sequences (e.g., see Gardes & Bruns 1993 paper describing amplification of basidiomycete ITS sequences from mycorrhiza samples). primer name ITS1 sequence (5'->3') comments TCCGTAGGTGAACCTGCGG reference White et al, 1990 ITS2 GCTGCGTTCTTCATCGATGC (is similar to 5.8S below) White et al, 1990 ITS3 GCATCGATGAAGAACGCAGC (is similar to 5.8SR below) White et al, 1990 ITS4 TCCTCCGCTTATTGATATGC White et al, 1990 ITS5 GGAAGTAAAAGTCGTAACAAGG (is similar to SR6R) White et al, 1990 ITS1-F CTTGGTCATTTAGAGGAAGTAA Gardes & Bruns, 1993 ITS4-B CAGGAGACTTGTACACGGTCCAG Gardes & Bruns, 1993 5.8S CGCTGCGTTCTTCATCG Vilgalys lab 5.8SR TCGATGAAGAACGCAGCG Vilgalys lab SR6R AAGWAAAAGTCGTAACAAGG Vilgalys lab back to the top of this page Intergenic spacer (IGS) primers (including 5S RNA primer sequences for basidiomycete fungi) The greatest amount sequence variation in rDNA exists within the IGS region (sometimes also known as the non-transcribed spacer or NTS region). The size of the IGS region may vary from 2 kb upwards. It is not unusual to find hypervariability for this region (necessitating cloning of individual repeat haplotypes). Several patterns of organization can be found in different groups of fungi: 1. Most filamentous ascomycetes have a single uninterrupted IGS region (between the end of the LSU and start of the next SSU sequence), which may vary in length from 2-5 kb or more. Amplification of the entire IGS region requires using primers anchored in the 3' end of the LSU gene (e.g., LR12R) and 5' end of the SSU RNA gene (e.g., invSR1R). 2. In many ascomycetous yeasts and nearly all basidiomycetes, the IGS also contains a single coding region for the 5S RNA gene, which divides the IGS into two smaller regions that may be more easily amplified using. Depending on the orientation and position of the 5S RNA gene, the PCR may be used to sequentially amplify either aportion of the intergenic spacer region (IGS) beyond the large subunit RNA coding region. primer name sequence (5'->3') comments reference LR12R GAACGCCTCTAAGTCAGAATCC located within the LSU RNA (see above) Vilgalys lab invSR1R ACTGGCAGAATCAACCAGGTA located within the SSU RNA (positions 21-1) Vilgalys lab 5SRNA ATCAGACGGGATGCGGT (complementary to 5S RNA positions 46-26) Vilgalys lab 5SRNAR ACQGCATCCCGTCTGAT (5S RNA positions 26-46) Vilgalys lab back to the top of this page REFERENCES Bruns, T. D., R. Vilgalys, S. M. Barns, D. Gonzalez, D. S. Hibbett, D. J. Lane, L. Simon, S. Stickel, T. M. Szaro, W. G. Weisburg, and M. L. Sogin. 1992. Evolutionary relationships within the fungi: analyses of nuclear small subunit rRNA sequences. Molec. Phylog. Evol. 1: 231-241. Bruns, T. D., T. J. White, and J. W. Taylor. 1991. Fungal molecular systematics. Ann. Rev. Ecol. Syst. 22: 525-564. DePriest, P. T., and M. D. Been. 1992. Numerous group I introns with variable distributions in the ribosomal DNA of a lichen fungus. J. Mol. Biol. 228: 315-321. Elwood, H. J., G. J. Olsen, and M. L. Sogin. 1985. The small subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustula. Mol. Biol. Evol. 2: 399-410. Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizae and rusts. Mol. Ecol. 2: 113-118. Gargas, A., and P.T. DePriest. 1996. A nomenclature for fungal PCR primers with examples from intron-containing SSU rDNA. Mycologia 88: 745-748 Gargas, A., and J.W. Taylor. 1992. Polymerase chain reaction (PCR) primers for amplifying and sequencing 18S rDNA from lichenized fungi. Mycologia 84: 589-592. Gerbi, S. A. 1986. Chapter 7 - Evolution of ribosomal DNA. Pp. 419-517 In: Molecular evolution, ed. McIntyre, R. Hibbett, D. S. 1991. Phylogenetic relationships of the Basidiomycete genus Lentinus: evidence from ribosomal RNA and morphology. Ph.D. Thesis, Duke University, 1991. Hibbett, D. S. 1992. Ribosomal RNA and fungal systematics. Trans. Mycol. Soc. Jpn. 33: 533-556. Hibbett, D. S., and R. Vilgalys. 1991. Evolutionary relationships of Lentinus to the Polyporaceae: evidence from restriction analysis of enzymatically amplified ribosomal DNA. Mycologia 83: 425439. Hibbett, D. S., and R. Vilgalys. 1993. Phylogenetic relationships of the Basidiomycete genus Lentinus inferred from molecular and morphological characters. Syst. Bot. 18: 409-433. Hillis, D. M., and M. T. Dixon. 1991. Ribosomal DNA: molecular evoluiton and phylogenetic inference. Quart. Rev. Biol. 66: 411-453. Hopple, J. S., Jr., and R. Vilgalys. 1994. Phylogenetic relationship among coprinoid taxa and allies based on data from restriction site mapping of nuclear rDNA. Mycologia 86: 96-107. Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci., U. S. A. 82: 6955-6959. Vilgalys, R., and D. Gonzalez. 1990. Organization of ribosomal DNA in the basidiomycete Thanatephorus praticola. Curr. Genet. 18: 277-280. Vilgalys, R., J. S. Hopple, Jr., and D. S. Hibbett. 1994. Phylogenetic implications of generic concepts in fungal taxonomy: The impact of molecular systematic studies. Mycologica Helvetica 6: 73-91. White, T. J., T. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pp. 315-322 In: PCR Protocols: A Guide to Methods and Applications, eds. Innis, M. A., D. H. Gelfand, J. J. Sninsky, and T. J. White. Academic Press, Inc., New York. ©W°







