Methods
advertisement

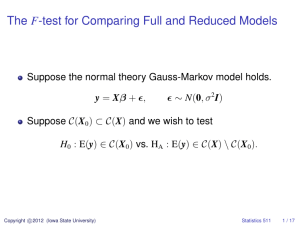
Supplemental Material Decreased nNOS within the PVN leads to Increased Sympatho-Excitation in Chronic Heart Failure: Role for Angiotensin II and CAPON. Neeru M Sharma1, Hong Zheng1, Parmender P. Mehta2, Yi-Fan Li3 & Kaushik P. Patel1 1 Department of Cellular and Integrative Physiology, University of Nebraska Medical Center, Omaha, NE 68198-5850; 2Department of Biochemistry and Molecular Biology, University of Nebraska Medical Center, Omaha, NE 68198-5850; 3Division of Basic Biomedical Science, College of Medicine, University of South Dakota, Vermillion, SD, 57069 Methods Immunofluorescent staining of PVN sections To evaluate the relationship between CAPON and nNOS in the PVN, sections were prepared for immunofluorescent staining. The rats were anesthetized with pentobarbital (65 mg/kg) and perfused transcardially with 150 ml of heparinized saline followed by 250 ml of 4% paraformaldehyde in 0.1 M sodium phosphate buffer. The brain was postfixed at 4°C for 4 h in 4% paraformaldehyde solution and then placed in 30% sucrose for 72 h. The brain was blocked in the coronal plane and sections 30 µm in thickness were cut with a cryostat. The free floating sections were incubated with 10% normal donkey serum, 0.02% Triton X in phosphate buffered saline (PBS) for 1 h at room temperature and then incubated with primary antibody against CAPON (rabbit antibody, 1:500, Santa Cruz Biotechnology, Inc, CA), and nNOS (mouse antibody, 1:1000, Santa Cruz Biotechnology, Inc, CA) overnight at 4°C. After washing with PBS, the sections were incubated with Cy3-conjugated donkey anti-rabbit (1:500, Jackson ImmunoResearch, PA) and Cy2-conjugated donkey anti-mouse secondary antibody (1:1000, Jackson ImmunoResearch, PA) for 4 h at room temperature. The nuclei were stained by DAPI (Molecular Probes, CA). After washing with PBS, the sections were mounted on slides and cover-slipped with fluoromounting-G (SouthernBiotech, AL). Distribution of immunofluorescence within the PVN was viewed using an Olympus fluorescence microscope (Japan) equipped with a digital camera (Qimaging, Canada) Real-time PCR Total RNA for real time PCR was isolated from PVN punches or cell line by Tri-Reagent (MRC) method as per the manufacturer’s instructions1. Briefly, the homogenate was separated into organic and aqueous phases by the addition of bromochloropropane and subsequent centrifugation. The RNA, contained in the aqueous phase, was precipitated with isopropanol, washed with ethanol, and solubilized in 10 l nuclease free water. Following extraction of mRNA, samples underwent reverse transcription for 40 min at 37C in the presence of 1.5 M random hexamers and 100 U MMLV-RTase. Real-time RT-PCR measurements were made using the iCycler iQ Multicolor Real-Time Detection System with output to a computer-based acquisition system (Bio-Rad). The protocol consisted of denaturation (95C for 3 min), amplification and quantification repeated 50 times (95C for 10 s, 55C for 45 s), denaturation at 95C for 1 min, reannealing at 55C for 1 min, and a melt curve (55-95C with a heating rate of 0.5C per 10 s). The reaction mixture consisted of SYBR Green Supermix (Bio-Rad), 10 nM forward primer, 10 nM reverse primer, nuclease free H2O, and the cDNA template of interest. Sequences of primers used for real time PCR were given in online data supplement Table 1. Relative expression of CAPON/nNOS was calculated using the 2-CT method, which relates expression of the target gene to expression of a reference gene2. Western blot To analyze proteins by immunoblotting sample preparation from cell/tissue was done as described previously.1 Briefly, protein was extracted from PVN punches of rat or cell samples in 100 l protein extraction buffer (10 mM Tris, 1 mM EDTA, 1% SDS, 0.1% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride). Protein content of lysate was estimated using the bicinchoninic acid method with bovine serum albumin as standard (Pierce, Rockford, IL). Samples were adjusted to contain the same concentration of total protein using 1% SDS, and then equal volumes of 2 X 4% SDS sample buffer were added. Protein lysates (20-30 g) were fractionated on 7.5% SDS-polyacrylamide gel in Tris-glycine running buffer. Proteins were electrophoretically transferred onto PVDF membranes at 300 mA for 90 min. Non-specific binding sites were blocked by incubating the membrane with 5% nonfat dry milk (w/v) in TBST (10 mM tris, 150 mM NaCl, 0.05% Tween-20) at ambient temperature for 1h. Membrane was incubated overnight either with rabbit polyclonal anti CAPON (0.4 g/ml) or mouse monoclonal nNOS (0.4 g/ml) from Santa Cruz at 40C, subsequently washed three times with TBST and then incubated with horseradish peroxidase conjugated anti-rabbit IgG (1:3333, Pierce) at room temperature for 1h. Protein bands were visualized by enhanced chemiluminescence substrate (Pierce), followed by an exposure within an Epi Chemi II darkroom (UVP BioImaging, Upland, CA) for visualization with the work lab digital imaging system. NIH Image J program was used to highlight the bands and quantify the signal. Coimmunoprecipitation: Cells were lysed 24 hr after transfection in lysis buffer. Lysates were cleared by centrifugation, and the supernatants normalized for protein content were subjected to preclearing with protein A/G agarose (Santa Cruz Biotechnology) for 30 min. Cleared supernatants (400ug) were incubated with CAPON antisera overnight at 40C. The immune complexes were precipitated after 2h incubation with protein A/G agarose by centrifugation. The precipitates were washed twice with icecold lysis buffer. For western blotting, samples were dissolved in sample buffer (200 mMTris-HCl, pH 6.8, 6% SDS, 20% glycerol, 10% DTT, and 0.1 mg/ml bromophenol blue), separated by SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Immunofluorescent microscopy of NG108 cells Adherent NG108 cells were grown on laminin coated 6mm Transwel-ClearTM inserts (Corning, Costar) overnight. Cells were fixed in 4% paraformaldehyde in PBS for 10 min and then permeabilized with 0.2% Triton X-100 for 20-30 min. Normal goat serum (10%) was used for blocking for 1 h followed by incubation with primary antibody at 4C overnight. All antibodies used were diluted in 1% blocking solution. For nNOS (mouse antibody, Santa Cruz Biotechnology, Inc, CA) and CAPON (rabbit antibody, Santa Cruz Biotechnology, Inc, CA) double immunostaining, dilutions of 1:100 for mouse anti-nNOS and 1:200 for rabbit anti-CAPON were used. Neurons were then washed with PBS three times. Secondary antibody consisted of a 1:200 dilution of Cy2-conjugated donkey antimouse IgG and Cy3-conjugated donkey anti-rabbit was used for 2h in dark. Coverslips were then mounted onto frosted glass microscope slides using Fluoromount G (Southern Biotechnology). Labeled cells were visualized by Olympus fluorescence microscope equipped with digital camera Table 1: Primers used for Real time PCR. Gene Reference Forward primer Reverse primer sequence* RPL19 NM_031103 5’-CCCCAATGAAACCAACGAAA -3’ 5’-ATGGACAGTCACAGGCTTC -3’ nNOS NM_052799 5’- ATCACAAGCCTATGCCAAGACC -3’ 5’-TCTCCATTGCCAAAGGTGCTG -3’ 5’-GGAGCTCATCAAGTTCCGAGT-3’ 5’- TCATCCAAACTGTCACCCAA -3’ CAPON NM_138922 *Accession numbers in NCBI GeneBank for the sequence used in designing the primers References 1. 2. Kleiber AC, Zheng H, Sharma NM, Patel KP. Chronic AT 1 receptor blockade normalizes NMDA-mediated changes in renal sympathetic nerve activity and NR1 expression within the PVN in rats with heart failure. Am J Physiol Heart Circ Physiol 2010;1152:1546-1555. Livak JK, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2-ddCt method. Methods 2001;25:402-408.