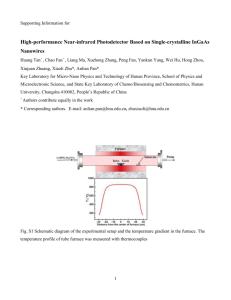
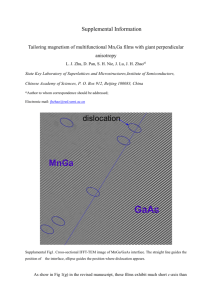
Supporting materials
advertisement

Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice Li Zhang, Qiuping Li, Haijiao Dong, Qin He, Liwen Liang, Cong Tan, Zhongmin Han Wen Yao, Guangwei Li, Hu Zhao, Weibo Xie, Yongzhong Xing* Supporting materials Table S1 Information of 107 wild rice accessions used in this study Accession Sources Accession Sources Accession Sources S1001 GAAS 11SW5 S1007 GAAS 11SW6 GAAS IRGC104833 IRRI GAAS IRGC105568 IRRI S1016 GAAS 11SW7 GAAS IRGC105569 IRRI S1020 GAAS 11SW8 GAAS IRGC105735 IRRI S1041 GAAS 11SW9 GAAS IRGC105956 IRRI S1054 GAAS 11SW10 GAAS IRGC81886 IRRI S1062 GAAS 11SW11 GAAS IRGC81900 IRRI S1064 GAAS 11SW12 GAAS IRGC81989 IRRI S1067 GAAS 11SW13 GAAS IRGC93216 IRRI S1069 GAAS 11SW14 GAAS IRGC104599 IRRI S1081 GAAS 11SW15 GAAS IRGC104709 IRRI S1084 GAAS dongyezhangtang-7 GAAS 07YR09 HZAU S1091 GAAS dongyezhangtang-6 GAAS CLR HZAU S1098 GAAS dongyedongtang-1 GAAS S2032 HZAU S1102 GAAS dongyeshuitaoshuixia-1 GAAS IRGC80643 IRRI S1107 GAAS Linchanganjiashan-2 GAAS IRGC80752 IRRI S1112 GAAS dongyeyechansi-2 GAAS IRGC81985 IRRI S1124 GAAS dongyekanxialong-3 GAAS IRGC83795 IRRI S1133 GAAS dongyuanlinchang-1 GAAS IRGC103844 IRRI S1141 GAAS dongyedongtangshang-1 GAAS IRGC104624 IRRI S1143 GAAS dongyedongtangxia-2 GAAS IRGC104647 IRRI S1160 GAAS Y079 HAAS IRGC105314 IRRI S1212 GAAS Y117 HAAS IRGC105349 IRRI S1216 GAAS Y118 HAAS IRGC105375 IRRI S1192 GAAS Y120 HAAS IRGC105422 IRRI S1204 GAAS Y131 HAAS IRGC105569 IRRI S1109 GAAS Y149 HAAS IRGC105709 IRRI S03005 GAAS Y182 HAAS IRGC105738 IRRI S03013 GAAS Y190 HAAS IRGC105829 IRRI S03035 GAAS Y204 HAAS IRGC105887 IRRI S03038 GAAS Y279 HAAS IRGC105960 IRRI S03045 GAAS IRGC81984 IRRI IRGC106078 IRRI 11SW1 GAAS IRGC100916 IRRI IRGC106424 IRRI 11SW2 GAAS IRGC103308 IRRI IRGC106452 IRRI 11SW3 GAAS IRGC103423 IRRI YSD7 JAAS 11SW4 GAAS IRGC104501 IRRI GAAS: Guangdong Academy of Agricultural Sciences; HAAS: Hunan Academy of Agricultural Sciences; HZAU: Huazhong Agricultural university; IRRI: International Rice Research Institute; JAAS: Jiangxi Academy of Agricultural Sciences. Table S2 List of 41 CCT domain genes identified in rice CCT-ID OsCCT01 OsCCT02 OsCCT03 OsCCT04 OsCCT05 OsCCT06 OsCCT07 OsCCT08 OsCCT09 OsCCT10 OsCCT11 OsCCT12 OsCCT13 OsCCT14 OsCCT15 OsCCT16 OsCCT17 OsCCT18 OsCCT19 OsCCT20 OsCCT21 Chr 1 2 2 2 2 2 2 2 2 3 3 3 3 3 3 4 5 5 6 6 6 LOC Os01g61900 Os02g01990 Os02g05470 Os02g05510* Os02g08150 Os02g39710 Os02g40510 Os02g49230 Os02g49880 Os03g04620 Os03g17570 Os03g22770 Os03g47970* Os03g50310 Os03g52450* Os04g42020 Os05g38990 Os05g51690 Os06g01340 Os06g15330 Os06g16370 SubC CMF CMF CMF CMF COL COL PRR COL COL CMF PRR COL CMF COL CMF COL CMF CMF COL COL COL CCT-ID OsCCT22 OsCCT23 OsCCT24 OsCCT25 OsCCT26 OsCCT27 OsCCT28 OsCCT29 OsCCT30 OsCCT31 OsCCT32 OsCCT33 OsCCT34 OsCCT35 OsCCT36 OsCCT37 OsCCT38 OsCCT39 OsCCT40 OsCCT41 Chr 6 6 6 6 7 7 7 8 8 9 9 9 10 10 11 11 11 12 12 12 *These 6 genes did not reported by Cockram et al. (2012). LOC Os06g19444 Os06g44450 Os06g48534* Os06g48610 Os07g15770 Os07g47140 Os07g49460 Os08g15050 Os08g42440 Os09g06464 Os09g33550 Os09g36220 Os10g32900 Os10g41100 Os11g01074.4 Os11g01100* Os11g05930 Os12g01080 Os12g01100* Os12g16160 SubC COL COL CMF CMF CMF COL PRR COL COL COL COL PRR CMF CMF CMF CMF PRR CMF CMF CMF Table S3 Primers used for amplifying gene fragments and transcriptional expression analysis Gene PCR Forward primer Reverse primer OsCCT01 OsCCT01 OsCCT03 OsCCT03 OsCCT05 OsCCT07 OsCCT07 OsCCT09 OsCCT10 OsCCT11 OsCCT17 OsCCT18 OsCCT19 OsCCT19 OsCCT22 OsCCT22 OsCCT29 OsCCT33 OsCCT37 OsCCT38 OsCCT39 OsCCT39 OsCCT40 OsCCT41 OsCCT01 OsCCT01 Hd1 Ehd1 Hd3a RFT1 OsCCT01 OX RNAi OX RNAi RNAi OX RNAi OX OX RNAi RNAi OX OX RNAi OX RNAi RNAi RNAi OX RNAi OX RNAi OX RNAi SC EP qPCR qPCR qPCR qPCR qPCR ggtacctgaccctaccacctctcacag ggactagtggtaccctgttgtgcaggaaatgttac cccgggtctgaaaggcgttctgctga aaaactagtggtaccaaggtttgttaggagcccatc actagtggtaccaaccagatgtcctcgtcga ggtaccctgtgctgttcgtgctgatt actagtggtaccacttgcatctggagcgaagc ggtaccagatcgagtgcaagtgagctgc ggtacccgatatctcacgcccatact ggactagtggtatttcaggtggccataatgga actagtggtaccagatagcaggccaagggt ggtacctccactgtctccgttcattg ggatccgggtaatagtcgagaagggtt aaactagtggtaccttcagcaagcagatcaagtatgc aacgggtacctgcattcagctaatgctctttctcg actagtggtaccggtatgcatctcgcaaggct actagtggtaccaccgcaagttccagaagacca actagtggtaccttgatgggcagccattctgg ggatcctgtggacctttagattgc actagtggtaccctgctataccctaccactacg ggatcctgttgggctgatgtatgt actagtggtaccgacgaataataatgtgccggaggat cccttcctcctcactcccactg ggactagtggtacctatgcttgccggaagacgctc tctagaatgttccgccattcctcctctg ggatccatcacctgttctagtcacac tcagcaacagcatatctttctcatca tggaaatctcgaaaaacccg gctcactatcatcatccagcatg tgacctagattcaaagtctaatcctt tacctcgacggcaatgttag ggatccccactctgacctgacctgac cgagctcggatccgattaattacctctgctcgc tctagacgtagtttccaggcgagtt aaagagctcggatccgcacattgtaaacctgagac gagctcggatccgctaatcctatgcatggtga gtcgacctaatgagttgcaaagtgagcag gagctcggatccccgttcagaccgactacatg gtcgacgagtccatgaggcatacgatcg ggatcccacttctcttgcgaatagag cgagctcggatcttcactggttcgatcctggt gagctcggatcccgaagttacactacggcga ggatcctcactggcagctcgtcctct gtcgacaaggtccagaagatgaaagaat aaagagctcggatccgataggagtatcaatgagc aacggtcgacttgatgcttgctaacctaacacttgg gagctcggatcccacagcaagggttggtggat gagctcggatccaccattagcatgacggcga gagctcggatccggttggaagagctgtcacag ctgcagtgttgctcacgcttgtat gagctcggatcctactgcagttctggacaccg aagcttaggtgaccgtctatgaaa gagctcggatccgagggtgtaaattgaaccgtctgca ggccgatctagatcattgagg cgagctcggatccatcctagcagctactacgaatc tctagacgaggcgaggttcacggacag aagctttgtgcgctggcttatggtg tctggaatttggcatatctatcacc gcgctagcaaagcttcggt ccttgctcagctatttaattgcataa tgccggccatgtcaaattaataac ctgatgctcctctggatgtt OX and RNAi, primers used to amplify the gene fragmentsfor overexpression and RNA interference,respectively.SCprimers used for protein subcellular localization,EP primers used for gene expression patterns. Table S4 Transformation information of 18CCT genes CCT-ID OsCCT01 OsCCT03 OsCCT05 OsCCT07 OsCCT09 OsCCT10 OsCCT11 OsCCT17 OsCCT18 OsCCT19 OsCCT22 OsCCT29 OsCCT33 OsCCT37 OsCCT38 OsCCT39 OsCCT40 OsCCT41 Donor Minghui63 Nipponbare Minghui63 Minghui 63 Minghui63 Nipponbare Minghui63 Nipponbare Nipponbare Nipponbare Minghui63 Minghui63 Nipponbare Minghui63 Nipponbare Minghui63 Nipponbare Nipponbare Receptor Zhonghua 11 Hejiang19 Zhonghua 11 Zhonghua 11 Zhonghua 11 Zhonghua 11 Zhonghua 11 Zhonghua 11 Hejiang19 Hejiang19 Zhonghua 11 Zhonghua 11 Zhonghua 11 Zhonghua 11 Zhonghua 11 Zhonghua 11 Zhonghua 11 Zhonghua 11 Vector 1301s,ds1301 1301s,ds1301 ds1301 1301s, ds1301 1301s 1301s ds1301 ds1301 1301s 1301s,ds1301 1301s,ds1301 ds1301 ds1301 1301s ds1301 1301s,ds1301 1301s ds1301 Gene fragments DNA CDS DNA DNA DNA DNA DNA DNA DNA DNA CDS DNA CDS DNA CDS DNA DNA DNA Table S5 List of 59 heading date QTLs identified in rice QTL-I D Qhd01 Qhd02 Qhd03 Qhd04 Qhd05 Qhd06 Qhd07 Qhd08 Qhd09 Qhd10 Qhd11 Qhd12 Qhd13 Qhd14 Qhd15 Qhd16 Qhd17 Qhd18 Qhd19 Qhd20 Qhd21 Qhd22 Qhd23 Qhd24 Qhd25 Qhd26 Qhd27 Qhd28 Qhd29 Qhd30 QTLs Chr. Reference QHd1a QHd1b AQEA470 AQFW073 qDTH-1 AQEA472 hd1 Hd1a dth2.1 QTL2a AQEA490 QTL2b AQEA506 DTH2 QHd2b Hd9 Hd8 hd3 QHd3b Hd6 dth4.1 AQEA499 dth4.2 AQFW133 QHd5a QAFW190 qHD-5 QHd5b Hd6a Hd6b 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 3 3 3 3 3 4 4 4 4 5 5 5 5 6 6 1 3 1 3 4 1 6 3 8 5 1 5 1 11 1 12 14 16 3 17 18 1 18 3 1 3 21 1 23 23 QTL-I D Qhd31 Qhd32 Qhd33 Qhd34 Qhd35 Qhd36 Qhd37 Qhd38 Qhd39 Qhd40 Qhd41 Qhd42 Qhd43 Qhd44 Qhd45 Qhd46 Qhd47 Qhd48 Qhd49 Qhd50 Qhd51 Qhd52 Qhd53 Qhd54 Qhd55 Qhd56 Qhd57 Qhd58 Qhd59 QTLs Chr. Reference Hd1 AQFW132 AQEA572 AQEA469 QTL7a Ghd7 AQFW072 Hd2 Ghd7.1 Ghd8 QHd8 QHd8a qDTH-8 QHd8b QHd9 qHDD9-1 qDTH-9 qDTH-10 Hd-10b qQTL-10b AQAT006 AQEA601 QHd11 Dth11 hd11 AQEA607 dth12.1 QHd12 AQFW196 6 6 6 6 7 7 7 7 7 8 8 8 8 8 9 9 9 10 10 10 10 11 11 11 11 11 12 12 12 2 3 1 1 5 2 3 7 9 10 3 1 4 1 1 13 15 4 6 5 19 1 3 20 16 1 22 1 3 References 1. Li, Z. et al. QTL× environment interactions in rice. I. Heading date and plant height. Theoretical and Applied Genetics 108, 141-153 (2003). 2. Yano, M. et al. Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theoretical and Applied Genetics 95, 1025-1032 (1997). 3. Mei, H. et al. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations. Theoretical and Applied Genetics 110, 649-659 (2005). 4. Cho, Y. et al. QTLs analysis of yield and its related traits in wild rice relative Oryza rufipogon. Treat Crop Res 4, 19-29 (2003). 5. Zhou, Y. et al. Genetic dissection of heading time and its components in rice. Theoretical and Applied Genetics 102, 1236-1242 (2001). 6. Lu, C. et al. Comparative mapping of QTLs for agronomic traits of rice across environments by using a doubled-haploid population. Theoretical and applied genetics 94, 145-150 (1997). 7. Yamamoto, T., Lin, H., Sasaki, T. & Yano, M. Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny. Genetics 154, 885-891 (2000). 8. Septiningsih, E. et al. Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theoretical and Applied Genetics 107, 1419-1432 (2003). 9. Yamamoto, T., Kuboki, Y., Lin, S., Sasaki, T. & Yano, M. Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factors. Theoretical and Applied Genetics 97, 37-44 (1998). 10. Yan, W.-H. et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant 4, 319-330 (2011). 11. Wu, W. et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proceedings of the National Academy of Sciences 110, 2775-2780 (2013). 12. Lin, H., Ashikari, M., Yamanouchi, U., Sasaki, T. & Yano, M. Identification and characterization of a quantitative trait locus, Hd9, controlling heading date in rice. Breeding Science 52, 35-41 (2002). 13. Hittalmani, S. et al. Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia. Theoretical and Applied Genetics 107, 679-690 (2003). 14. Takeuchi, Y., Lin, S., Sasaki, T. & Yano, M. Fine linkage mapping enables dissection of closely linked quantitative trait loci for seed dormancy and heading in rice. Theoretical and Applied Genetics 107, 1174-1180 (2003). 15. SangNag, A., YoungChan, C., KyongHo, K., ImSoo, C. & YeonGyu, K. Mapping of QTLs for yield traits using an advanced backcross population from a cross between Oryza sativa and O. glaberrima. Korean Journal of Breeding 37, 214-220 (2005). 16. Yu, S. et al. Identification of quantitative trait loci and epistatic interactions for plant height and heading date in rice. Theoretical and Applied Genetics 104, 619-625 (2002). 17. Takahashi, Y., Shomura, A., Sasaki, T. & Yano, M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proceedings of the National Academy of Sciences 98, 7922-7927 (2001). 18. Thomson, M. et al. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theoretical and Applied Genetics 107, 479-493 (2003). 19. Wang, C., Yasui, H., Yoshimura, A., Wan, J. & Zhai, H. Identification of quantitative trait loci controlling F2 sterility and heading date in rice]. Yi chuan xue bao 29, 339 (2002). 20. Xiao, J., Li, J., Yuan, L. & Tanksley, S. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theoretical and Applied Genetics 92, 230-244 (1996). 21. Zou, J. et al. Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.). Theoretical and Applied Genetics 101, 569-573 (2000). 22. Xiao, J. et al. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150, 899-909 (1998). 23. Xing, Y., Xu, C., Hua, J., Tan, Y. & Sun, X. Mapping and isolation of quantitative trait loci controlling plant height and heading date in rice. Acta Botanica Sinica 43, 721-726 (2000). Table S6 The genomic nucleotide diversity of CCT family 107common wild rice accessions CCT-ID SNP π (10-3) πc/πw Θ (10-3) sites c w c w OsCCT01 42 2.9 2.9 0.99 1.3 2.0 OsCCT02 67 2.1 3.2 0.66 2.3 2.9 OsCCT03 42 1.3 0.9 1.44 0.9 1.2 OsCCT04 66 2.0 1.6 1.17 1.2 1.8 OsCCT05 82 2.9 3.6 0.81 2.0 3.6 OsCCT06 76 3.4 2.6 1.31 2.2 3.3 OsCCT07 123 2.7 2.9 0.93 2.7 3.2 OsCCT08 137 2.6 2.7 0.96 1.5 2.3 OsCCT09 87 2.1 2.0 1.10 1.2 2.4 OsCCT10 77 1.1 1.6 0.69 0.9 1.4 OsCCT11 211 1.6 2.5 0.64 1.3 2.4 OsCCT12 150 2.0 4.9 0.41 3.0 5.4 OsCCT13 89 2.5 2.4 1.04 1.5 2.2 OsCCT14 95 2.5 3.8 0.66 2.0 3.2 OsCCT15 77 1.8 1.4 1.29 1.0 1.5 OsCCT16 75 2.2 4.5 0.49 1.3 3.6 OsCCT17 34 1.5 1.7 0.84 0.5 1.2 OsCCT18 94 0.1 2.6 0.04 1.1 2.3 OsCCT19 61 3.5 2.8 1.25 2.0 3.2 OsCCT20 82 6.0 4.1 1.46 2.1 3.3 OsCCT21 105 2.3 2.3 0.99 1.2 3.8 OsCCT22 96 2.6 2.3 1.09 1.4 1.9 OsCCT23 49 2.6 2.5 1.04 1.5 2.0 OsCCT24 92 3.7 3.4 1.09 1.6 2.2 OsCCT25 146 3.6 2.5 1.44 2.2 3.7 OsCCT26 165 6.0 5.5 1.09 3.2 5.4 OsCCT27 129 2.2 4.0 0.55 1.5 4.1 OsCCT28 199 1.9 1.8 1.06 1.1 1.7 OsCCT29 150 7.8 8.4 0.93 5.7 7.1 OsCCT30 92 1.9 1.8 1.06 1.1 1.7 OsCCT31 421 3.0 3.2 0.94 1.9 3.1 OsCCT32 80 2.5 1.5 1.67 1.7 2.0 OsCCT33 123 1.9 2.4 0.79 1.5 2.5 OsCCT34 55 2.5 1.9 1.32 1.3 1.9 OsCCT35 72 2.4 2.7 0.89 1.2 2.5 OsCCT36 10 0.3 0.2 1.50 0.2 0.2 OsCCT37 20 2.6 2.1 1.24 1.6 1.8 OsCCT38 121 2.3 2.4 0.96 1.5 2.4 OsCCT40 22 2.7 2.3 1.17 1.8 2.2 OsCCT41 88 4.8 3.1 1.55 3.3 3.8 genes in 529 cultivars and Tajima’s D c w 2.89* 1.20 -0.29 0.33 1.28 -0.85 1.74 -0.38 1.22 0.21 1.42 -0.69 -0.03 -0.24 2.03 0.52 1.94 -0.50 1.29 0.47 0.79 0.05 -0.95 -0.28 1.87 0.32 0.56 0.59 2.11 -0.28 1.63 0.72 3.89** 1.05 -2.31* 0.47 1.91 -0.39 4.97** 0.68 2.42 -1.25 2.14 0.68 1.96 0.79 3.66* 1.62 1.90 -1.01 2.52 0.01 1.35 -1.01 2.02 0.23 1.05 0.57 2.16 0.06 1.66 0.12 1.24 -0.80 0.80 -0.12 2.42 -0.03 2.36 0.20 2.03 0.56 1.44 0.40 1.46 0.03 1.10 0.18 1.28 -0.59 π, average number of nucleotide differences per site between two sequences; θ, Watterson estimator; Tajima’s D, test for neutral selection; *and ** Significant at P<0.01 and P<0.001 respectively. πc and πw indicate π in cultivars and π in wild rice respectively. Table S7 The nucleotide diversity of noncoding regions around OsCCT18 in cultivars and wild rice PositionS (bp) 29564885 29623975 29633070 29644549 29654549 29667564 29680439 29700011 29710059 Regions IG IT IT IG OsCCT18 IG IT IG IG N 529 529 529 529 529 529 529 529 529 Cultivars L S 2055 30 313 4 409 3 948 27 5726 45 805 13 906 5 1995 38 2013 18 H 68 7 5 15 27 11 6 45 27 N 107 107 107 107 107 107 107 107 107 Wild rice L S 2055 39 313 5 409 4 948 47 5726 71 805 24 906 8 1995 58 2013 22 πc/πw H 103 11 6 103 166 35 10 133 95 0.83 0.33 0.14 0.08 0.04 0.18 0.07 0.11 0.10 N: sample size; L: the length in bp from the first SNP to the last SNP; S: number of segregating sites; H: number of haplotypes; π: average proportion of pairwise differences per base pair. IG: intergenic region; IT: intron; πc and πw indicate π in cultivars and π in wild rice. Table S8 Phenotypes of OsCCT1 transgenic plants in the T2 generation under nature long day conditions in 2012 summer (Wu Han) Genotype Wild type Plant height (cm) Spikelets per panicle 100.7±3.8 239.3±24.8 Seed setting rate Length of internodes (cm) 1st 2nd 3rd 87.0±8.3 37.9±1.1 14.3±0 14.9±1.9 OXOsCCT1+ 69.3± 4.5 140.7±10.2 20.1± 25.0±0.9 .7 ± 9.5 6.2 ± 0.8 + +) OsCCT1RNAi 98.4±1.6 220.3±20.3 6.7 90.1±7.5 36.5±1.7 0.5 13.2±1 12.1±1.8 Data are present in means ± s.d. (N=15). The internodes indicate 1st.3 , 2nd and 3rd upmost stem internodes in the main culms. Figure S1 Population structure for cultivars in the study. Neighbour-joining tree of cultiva accessions, which was calculated from ~200000 SNPs randomly selected from the whole genome. Black, Ind; red, IndI; green, IndII ; yellow, TrJ; blue, TeJ; grey, Jap. Figure S2 Expression levels of 16 OsCCT genes in wild type (WT), negative transgenic plant (NP) and positive transgenic plant (PP) plants. Relative expression level of each gene compared with that of rice UBQ. Error bars indicate standard deviations, based on 3 biological replicates OsCCT-ZH11-OX represented Zhonghua 11 plants overexpressed OsCCT gene. OsCCT-ZH11-RNAi represented Zhonghua 11 plants suppressed OsCCT gene by RNAi. Figure S3 Expression levels of OsCCT01, 11 and 19 in wild type (WT), negative transgenic plant (NP) and positive transgenic plant (PP) plants. Relative expression level of each gene was compared with that of rice UBQ. Error bars indicate standard deviations, based on 3 biological replicates. HD means heading date scored as days to heading from sowing. OsCCT01-ZH11-OX, the genotype Zhonghua 11 overexpressed OsCCT01. OsCCT11-ZH11-RNAi, the genotype Zhonghua 11 suppressed OsCCT11 by RNAi. OsCCT19-HJ19-OX, the genotype Hejiang 19 overexpressed OsCCT19. Figure S4 Diurnal expression patterns of OsCCT01 in Zhonghua 11 Diurnal expression patterns of the OsCCT01 in Zhonghua 11 as indicated by quantitative RT-PCR results. In all panels the mean of each point is based on the average of two biological repeats calculated using the relative quantification method. The black bars indicate the dark period, and the white bars indicate the light period. The blue line indicates the long-day condition. The red line indicates the short-day condition. The numbers below the bars indicate hours of the day. Error bars indicate standard deviations. Figure S5 Subcellular localization of OsCCT01 Rice protoplasts were co-transformed with the fusion constructs 35s::GHD7:CFP (a) and 35s::OsCCT01:YFP (b); (c) bright field image and (d) merged image. Bar = 12 µm Figure S6 Diurnal expression patterns of OsGI, RID1 and Ehd3 in OsCCT01-positive transgenic plants and wild-type Zhonghua 11 plants grown under long day-length conditions and short day-length conditions X axis indicated the time points for sampling. Y axis indicated the relative expression levels of investigated genes to UBQ. Figure S7 Diurnal expression patterns of Ehd4, OsMADS50, OsMADS56, Ghd7, Ghd8, Ghd7.1 and DTH2 in OsCCT01-positive transgenic plants and wild-type Zhonghua 11 plants grown under long day-length conditions, Ehd4, OsMADS51 under short day-length conditions X axis indicated the time points for sampling. Y axis indicated the relative expression levels of investigated genes to UBQ.