Genetic diversity
advertisement

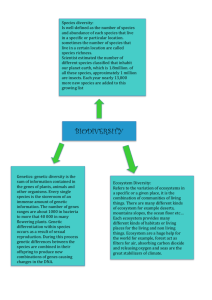
Genetic diversity Chapter 5 Genetic diversity occurs at 4 levels: Among species Among populations Within populations Within individuals Diversity between species Diversity within species Diversity within populations Diversity within individuals Measuring genetic diversity Protein electrophoresis (indirect method) Restriction Fragment Length Polymorphism (RFLP) Random Amplification of Polymorphic DNA (RAPDs) Microsatellites or Simple Sequence Repeat (SSR) Polymorphisms Amplified Fragment Length Polymorphisms(AFLP) DNA sequencing (direct method) Genetic variation in proteins Enzymes are run through electrophoresis Electrophoretically distinguishable forms of an enzyme are called allozymes Not all amino acid substitutions alter electrophoretic mobility Allozyme gel Table 5.1 Polymorphism (P) Proportion of genes that are polymorphic (frequency of common allele is less than 95%) Table 5.1 – Bison example 24 different genes, only 1 polymorphic MDH -1 has gene frequency of 0.6 for one allele and 0.4 for second allele Estimated polymorphism is 1/24 = 4.2% Heterozygosity (H) Proportion of genes at which the average individual is heterozygous In Bison example, 2/5 individuals were heterozygous, so H = 0.4 for this gene Sum heterozygosity across all genes and H = 0.4/24 = 0.017 Use of H Compare actual heterozygosity versus expected heterozygosity Partition variability to determine how much variability is due to variability among populations (Dst) and between populations (Hs) Ht = Hs + Dst Dst is related to Fst FST Which will have a higher Dst/Fst? Quantitative variation Continuous characters are polygenic and environmentally affected Complicates the picture Why is genetic diversity important? Evolutionary potential Loss of fitness Utilitarian values Evolutionary potential Genetic variation is the raw material for natural selection Greater diversity means greater ability to evolve with changing conditions Humans are top agent of change Selection by overharvesting Greater diversity means greater ability to colonize wider range of habitats Greater genetic diversity in amphibians living in forests than in aquatic habitats Genetic diversity is related to reproductive fitness Loss of fitness Inbreeding depression Isolated populations have more inbreeding Illinois prairie chicken Disease susceptibility in California sea lions Inbreeding increases extinction risk Heterosis Heterozygote advantage or hybrid vigor Evolutionary potential of heterozygous population A population dominated by heterozygotes will also have greater genetic variation Anomaly – Outbreeding depression Local adaptation Gene flow can reduce adaptiveness Eichornia paniculata Outcrossing Brazilian population and self-fertilizing Jamaican population Inbred each for 5 generations then outcrossed the inbred lines No evidence of heterosis in self-fertilizing population Utilitarian values Importance in domestic species Crop diversity What causes reduction in diversity? Small populations are prone to loss of genetic diversity through genetic drift Loss of heterozygosity in small populations Loss of heterozygosity (H) Genetic drift Random fluctuation in allele frequencies over time by chance Important in small populations Founder effect Bottleneck effect Genetic drift When a few individuals remain in a population (or found a new population), genetic constitution depends on the genes of the small population A low number of individuals may lead to low genetic diversity Gene frequency may not represent that of the population that founders came from Bottleneck effect Drastic reduction in population and gene pool size Founder Effect Effect of drift when a small number of individuals start a new population Effect is pronounced on isolated islands Contribution of 21 founders to the captive Guam Rail population Table 5.2 Table 5.3 Genetic drift Random change in allele frequencies due to sampling error in small populations Mathematically, genetic drift represents a chronic bottleneck that results in repeated losses of variability and eventual fixation of loci Table 5.4 Regeneration of diversity Loss of heterozygosity (H) Regeneration of diversity Mutation takes over 100 generations to regenerate diversity at a single locus Genetically effective population size (Ne) Variation from idealized population 1:1 sex ratio Sexually-reproducing Non-overlapping generations Even distribution of progeny among females No selection No mutation Population size relevant for determining genetic effects OR the number of individuals contributing genes to the next generation Ne is typically 1/3 – 1/4 of Nc Reasons why Ne<Nc Unequal numbers of progeny Unequal breeding sex ratio Fluctuating population size Assortative mating Unequal numbers of progeny Unequal breeding sex ratio Fluctuating population size Fluctuating population size Inbreeding Mating of close relatives leads to: Reduced heterozygosity Reduced fecundity Increased mortality Estimate inbreeding coefficient (F) Gathering genetic information How do you get DNA samples without disturbing the animals?