MResMolecularNeuroscienceProjects
advertisement

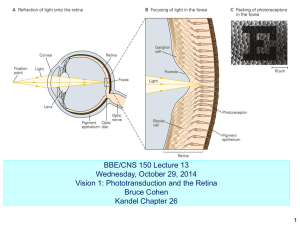
Projects on offer 1. The effect of Amyloid beta (Aβ) in neurotransmitter release Dr Pavlos Alifragis, School of Biological Sciences, Royal Holloway University London, Egham, Surrey, TW20 0EX. Pavlos.Alifragis@rhul.ac.uk The aim of this project is to investigate the effects of Aβ on neuronal and synaptic physiology. Alzheimer’s disease is strongly associated with Aβ deposits in the brain, and the soluble oligomeric forms of Aβ which correlate with cognitive decline. In certain brain regions, specifically the hippocampus, Aβ potently disrupts synaptic plasticity, by inhibiting the induction of LTP, and altering synaptic morphology. The mechanisms underlying this synaptic dysfunction remain largely unclear. Furthermore, the site of Aβ action remains elusive, with reports of toxic Aβ both internally and extracellularly, and evidence suggesting Aβ targets both the pre- and postsynapse. We have exciting data suggesting Aβ is interacting with presynaptic proteins such as Synaptophysin. Moreover we have shown that this interaction is necessary and sufficient to disrupt the interaction between Synaptophysin and VAMP2. In addition to the formation of intact synaptic vesicles, disrupting the interaction between VAMP2 and synaptophysin at the synapse affects the regulation of SNARE complex formation, as synaptophysin bound to VAMP2, prevents VAMP2 entering the SNARE complex allowing SV fusion and exocytosis. Our data show increased entry of VAMP2 into the SNARE complex, as a result of Aβ induced disruptions to the VAMP2-synaptophysin complex. The student will employ a range of techniques from culturing primary hippocampal neurons, to confocal microscopy, immuno labelling, treatment with Aβ etc. in order to investigate additional effects induced by Aβ at the presynaptic contact. The initial focus would be on Synapsin 1, a presynaptic protein involved in regulating the number of synaptic vesicles available for release via exocytosis. The project will investigate whether treatment of neurons with Aβ results in dissociation of Synapsin clusters at the presynaptic contact. References: 1) Hall, A.C., et al., Mol Cell Neurosci, 2002. 20(2): p. 257-70. 2. Characterisation of Smart Contrast Reagents for early Diagnosis of Alzheimer's Disease by MRI Prof Brian Austen, St. George’s University of London, Cranmer Terrace, London SW17 0RE. sghk200@sgul.ac.uk Early diagnosis of Alzheimer's at the mild cognitive impairment stage is essential if effective treatment and prevention is to be implemented. We have chemically synthesised several contrast agents, peptides that bind aggregated beta-amyloid, and contain ligated gadolinium(which enhances the T1 and suppresses the T2 modes of an MRI scan). The project will involve screening further reagents for binding to beta-amyloid dimers and fibrils using ELISA assays, and for potential toxicity to neuronal cells in culture, using biochemical tests. An understanding of methods presently used for imaging neurodegeneration will be obtained, along with knowledge of the main pathological mechanisms of Alzheimer's Disease. In collaboration with others in the lab, the student will examine the MRI changes resulting from the contrast agents injected into transgenic mice models of Alzheimer’s Disease. References: 1) Journal of Peptide Science 14 (8) 44 2) Brian M. Austen,‡ Katerina E. Paleologou,§ Sumaya A. E. Ali,| Mohamed M. Qureshi,| David Allsop,§ and Omar M. A. El-Agnaf, (2008) Designing Peptide Inhibitors for Oligomerization and Toxicity of Alzheimer’s /beta/-Amyloid Peptide. Biochemistry, 47(7):1984-92 3. Investigating the effects of the anticonvulsant, valproic acid on NMDA receptor function and pharmacology Dr Philip Chen, School of Biological Sciences, Royal Holloway University London, Egham, Surrey, TW20 0EX. philip.chen@rhul.ac.uk Fast neuronal communication at synaptic contacts relies on neurotransmitter release from the presynaptic terminal and the majority of excitatory neurotransmission within the mammalian central nervous system is mediated by the neurotransmitter glutamate. Glutamate acts on a number of ligand gated ion channels often referred to as 'ionotropic' receptors which when opened permit ion flux across the cell membrane and result in a change in neuronal excitability. One subtype of ionotropic glutamate receptor is the Nmethyl-D-aspartate receptor (NMDAR) and these receptors play a number of roles in normal and abnormal processes within the central nervous system (CNS), ranging from learning and memory to excitotoxic neuronal cell death following stroke or brain trauma. The involvement of NMDARs in such a broad range of CNS processes and disorders have triggered a great deal of interest in understanding the molecular determinants of NMDAR function. NMDARs are tetrameric protein complexes made up of different types of subunit (two NR1s and two NR2s, of which there are four subtypes; A, B, C and D) and the incorporation of different subunits within the NMDAR complex underlies the distinct functional properties seen amongst NMDAR subtypes. The following project will be offered in my laboratory. Valproic acid (VPA) is the most common therapy for epilepsy, however its influence on excitatory neurotransmitter function is unclear. The project will examine if valproic acid influences NMDA receptor (NMDAR) function and pharmacology in recombinant expression systems. We will examine the pharmacology of valproic acid by recording glutamate evoked inward currents from Xenopus oocytes injected with different NMDAR subtypes under two-electrode voltage clamp configuration. Work on this project may also be extended to examine the effects of valproic acid on native NMDAR function using extracellular electrode recordings from hippocampal slices and the effect of VPA on NMDAR triggered neuronal cell death in primary neuronal cultures. Methods used: Two-electrode voltage clamp electrophysiology, expression and functional characterisation of recombinant receptors in Xenopus oocytes, in vitro cRNA synthesis, Xenopus oocyte injection and incubation and primary neuronal cell culture. References: 1) Chen PE et al. Structural features of the glutamate binding site in recombinant NR1/NR2A N-methyl-D-aspartate receptors determined by site-directed mutagenesis and molecular modeling. (2005) Molecular Pharmacology. 67:1470-1484. 2) Chen PE and Wyllie DJA. Pharmacological insights obtained from structure-function studies of ionotropic glutamate receptors. (2006) British Journal of Pharmacology. 147: 839-853. 3) Terbach N and Williams RSB. Structurefunction studies for the panacea, valproic acid. (2009) Biochemical Society Transactions 37:112632. 4. Generating and expressing an HA-tagged Kinectin fusion protein Dr Jennifer Murdoch, School of Biological Sciences, Royal Holloway University London, Egham, Surrey, TW20 0EX. Jenny.murdoch@rhul.ac.uk Tulp3 has been identified as a novel negative regulator of the Sonic hedgehog signalling pathway, but the precise molecular function of Tulp3 is not known. Several putative interacting proteins have been identified from a yeast 2hybrid screen. This project will aim to investigate further the interaction with one of these proteins, Kinectin. The project will aim to clone the Kinectin cDNA into an appropriate expression vector, to generate and express an HA-tagged fusion protein. This project will initially involve bacterial transformation of an existing kinectin plasmid, to allow preparation of larger quantities of plasmid DNA. The kinectin cDNA will be subcloned from this vector into an alternative expression vector, using a PCR or restriction enzyme-based subcloning approach. The student will be expected to play a major role in designing an appropriate subcloning strategy. Correct subcloning will be confirmed by sequence analysis, using the appropriate bioinformatics tools. The HA-Kinectin construct will be transfected into mammalian cells (293T or MDCK cells) and the expression of tagged protein assessed by Western blotting. If time allows, the plasmid will be co-transfected with a construct for V5-tagged Tulp3. Immunoprecipitation with antiHA and anti-V5 antibodies will be performed. Evidence for Tulp3 and Kinectin coimmunoprecipitation will support the hypothesis that these proteins interact. References: 1) Patterson, V.L., Damrau, C., Paudyal, A., Reeve, B., Grimes, D.T., Stewart, M.E., Williams, D.J., Siggers, P., Greenfield, A. and Murdoch, J.N. (2009) Mouse hitchhiker mutants have spina bifida, dorso-ventral patterning defects and polydactyly: Identification of Tulp3 as a novel negative regulator of the Sonic hedgehog pathway. Human Molecular Genetics 18:17191739. 5. Molecular and pharmacological analysis of the psychoactive plant, Khat Dr. Jamal Nasir, St. George’s University of London, Cranmer Terrace, London SW17 0RE. jnasir@sgul.ac.uk AIMS: 1. Using in vivo pharmacological assays to identify the active components in Khat. 2. Using genomics technologies to generate a molecular profile of Khat. The psychoactive plant khat (Catha edulis), is widely used as a stimulant in parts of Africa and the Middle East. Although highly addictive and potentially dangerous, it remains legal in Britain, despite being banned across North America and most of Europe. Here, it has historically been widely used by the Somali community. However, its more widespread use has recently caused great concern and has been widely publicised in the media. Known to induce euphoria and improve mood, alertness, sexual potency and self confidence, it can also induce more severe symptoms, including sleeplessness and high blood pressure leading to cardiac arrest. It can also cause mental health problems involving paranoia and delusions and other symptoms resembling schizophrenia. These symptoms are generally attributed to amphetamine like compounds, but the biological properties of this plant are poorly understood at the molecular level. Further insights into the underlying biochemical pathways could yield important clues about the aetiology of schizophrenia. In collaboration with Professor Bramley (RHUL), Dr. James Moffatt and Hussein Mohamoud (SGUL), we intend to isolate extracts of this plant and undertake pharmacological and mass spectroscopy studies to identify active components of Khat. 6. The heparin/heparan sulphate binding properties of the BMP antagonist protein gremlin Dr Chris Rider, School of Biological Sciences, Royal Holloway University London, Egham, Surrey, TW20 0EX. c.rider@rhul.ac.uk The bone morphogenetic proteins have important roles in the development of the nervous system, as indeed in many other tissues. They are also up-regulated in adult CNS in response to injury and will undoubtedly influence cell fate at such locations. A full understanding of their activities will be important in developing new approaches to the treatment of nervous system injury. BMP activities are modulated by a number of selectivity antagonist proteins which block receptor binding. Some of these antagonists including noggin, cerberus and chordin were first recognised because of their pivotal roles in head and brain development. An increasing number of the BMPs are now known to bind to heparin and related polysaccharides found on cell surfaces and the extracellular matrix. This is also currently the case for the two of the antagonists, but we do not yet know how widespread this property is amongst this class of proteins. Such interactions will have the effect of restricting the diffusion of these proteins, giving rise to localised concentration gradients, which may be crucial in tissue morphogenesis (3). This project seeks to study the predicted heparin/heparin sulphate binding properties of a further BMP antagonist, gremlin. Using a heparin binding ELISA method (1) we have clearly established that commercially available recombinant human gremlin binds strongly to heparin. However one caveat is that this particular protein carries a poly-histidine tag, and since this tag is polybasic it is possible that it may generate an artefactual binding site for the highly acidic heparin-like molecules. We therefore need to confirm that gremlin without this tag still binds strongly. To this end the student will contribute to on-going efforts to express and purify a recombinant gremlin tagged instead with the peptide myc, using approaches we have previously employed for other proteins (2). The project will then progress to characterizing its heparin/heparan sulphate binding properties and cell culture studies aimed at elucidating the physiological importance of gremlin interactions with these polysaccharides. References: 1) S.M. Rickard, R. S. Mummery, B. Mulloy and C.C. Rider (2003) The Binding of Human Glial Cell-Line Derived Neurotrophic Factor (GDNF) to Heparin and Heparan Sulphate: Importance of 2O-Sulphate Groups and Effects on its Interaction with its Receptor GFRa1. Glycobiology 13, 419426. 2) I. Alfano, P. Vora, R.S. Mummery, B. Mulloy and C.C. Rider (2007) The Major Determinant of the Heparin Binding of Glial Cell-Line Derived Neurotrophic Factor is near the Aminoterminus and is Dispensable for Receptor Binding. Biochemical Journal 404, 131-140. 3) C. C. Rider and B. Mulloy (2010) The Bone Morphogenetic Protein and Growth Differentiation Cytokine Family, and their Protein Antagonists. Biochemical Journal 429, 1-12. 7. The role of valproic acid in Ca2+ signalling Dr Katalin Török, St. George’s University of London, Cranmer Terrace, London SW17 0RE. ktorok@sgul.ac.uk Valproic acid is a major drug used in the treatment of bipolar disorder and it has been established that it affects the metabolism of the Ca2+ releasing signalling molecule, IP3 (1). The downstream effects of how it further affects cell function, e.g. neuronal output, are not fully understood. The Ca2+ signalling pathway is instrumental in learning and responses of the limbic system to stimulation (2). The aim of this project is to determine how valproic acid, via IP3 modulates neuronal Ca2+ signalling. Plan of investigation: A. Ca2+ imaging: Primary neuronal cultures will be prepared from rat brain and its regions, e.g. hippocampus and the Ca2+ signalling pathway will be stimulated by glutamate and glutamate receptor agonists. Intracellular Ca2+ transients will be monitored by the fluorescent Ca2+ indicator fluo-4 and imaged by confocal microscopy. Valproic acid will be applied and the resulting spatio-temporal patterns of Ca2+ will be compared to the responses of untreated neurons. B. Imaging of the response of the Ca2+-activated regulatory protein calmodulin (CaM and the ‘memory molecule’ Ca2+.calmodulin-dependent protein kinase II (CaMKII). Neuronal cultures will be transfected with fluorescently tagged CaM and CaMKII and their functional mutants and the intracellular translocation of CaMKII will be studied. Outcome: A. The mutants of Ca2+ downstream signalling molecules CaM and CaMKII will provide novel insight into neuronal Ca2+ signal transduction mechanisms B. A novel understanding of how valproic acid affects Ca2+ and downstream Ca2+ signalling providing new insights into its mechanisms of action. This will be useful in considering novel drug targets for bipolar disorder. References: 1) R.S.B. Williams, L. Cheng, A. Mudge and A.J. Harwood (2002) A common mechanism of action for three mood-stabilizing drugs. Nature 417, 2925 2) P.A.A. Grant, S.L. Best, N. Sanmugalingam, R. Alessio, A.M. Jama, and K. Török A two-state model for Ca2+/CaM-dependent protein kinase II (CaMKII) in response to persistent Ca2+ stimulation in hippocampal neurons Cell Calcium (2008) 44, 465—478 8. Synapse to nucleus Ca2+ signalling and the effects of valproic acid Dr Katalin Török, St. George’s University of London, Cranmer Terrace, London SW17 0RE. ktorok@sgul.ac.uk The metabolism of the Ca2+ releasing signalling molecule, IP3 is affected in bipolar disorder and its treatment by valproic acid (1). The longer term manifestation of bipolar disorder and the effects of valproic acid depend on synapse to nucleus signalling resulting in changes in gene expression. Our hypothesis is that Ca2+ signals are transduced to the nucleus by the nuclear translocation of the Ca2+ binding regulatory protein, calmodulin (CaM). It has been established that CaM nuclear transport is facilitated and our hypothesis is that it is aided by Ca2+/CaMdependent protein kinases I (CaMKI) and (CaMKIV) (2,3). The aim of this project is to test this hypothesis and to determine how valproic acid modulates neuronal Ca2+ signalling from the synapse to the nucleus. Plan of investigation: A. CaM nuclear translocation. Primary neuronal cultures will be prepared from rat brain and its regions, e.g. hippocampus and the Ca2+ signalling pathway will be stimulated by glutamate and glutamate receptor agonists. CaM nuclear translocation monitored by already developed fluorescently tagged CaM and its functional muatants and imaged by confocal microscopy. Valproic acid will be applied and CaM nuclear translocation will be compared to the responses of untreated neurons. B. Imaging of the response and structure-function relationship of CaMKI and CaMKIV in CaM nuclear translocation and the effects of valproic acid. Neuronal cultures will be transfected with fluorescently tagged CaM and CaMKI and CaMKIV mutants to determine the functionally important portions of CaM and CaMKs in nuclear translocation. The effects of valproic acid will be tested. Outcome: A. Ca2+ signalling mechanism from the synapse to the nucleus by CaM and CaMKs will be better understood B. A novel understanding of how valproic acid affects Ca2 signalling to the nucleus will be obtained. This will be useful in considering novel drug targets for bipolar disorder. References: 1) R.S.B. Williams, L. Cheng, A. Mudge and A.J. Harwood (2002) A common mechanism of action for three mood-sabilizing drugs. Nature 417, 292-5 2) K. Török PERSPECTIVE: The regulation of nuclear membrane permeability by Ca2+ signaling: A tightly regulated pore or a floodgate? Science STKE 386 (2007) pe24. 3) R. Thorogate and K. Török Ca2+-dependent and independent mechanisms of calmodulin nuclear translocation. J. Cell Science 117 (2004) 5923-5936. 9. Molecular analysis of the role of the centrosome in neurogenesis Dr Chris Wilkinson, School of Biological Sciences, Royal Holloway University London, Egham, Surrey, TW20 0EX. Christopher.wilkinson@rhul.ac.uk Genetic defects in centrosome proteins are linked to a number of developmental diseases in which the brain is affected. The disease microcephaly, in which the size of the brain is considerably reduced but its normal architecture retained, is caused in several cases by mutations in genes that encode centrosome proteins. This is the organelle that organises the microtubule network and has important roles in forming the mitotic spindle. The mechanism by which these defects in this tiny organelle have such a dramatic effect on brain development is still not known. This project will seek to investigate the role of the centrosome in early embryonic neurogenesis by depleting these proteins and observing the effect on brain development. Early zebrafish embryos will be used as a model as their rapid development and transparency make them accessible to molecular and microscopic analyses, including time-lapse confocal microscopy of living embryos. This project will seek to model microcephaly by making zebrafish embryos with small brains. The challenge will then be to work out the molecular mechanisms underpinning the gross phenotype. Techniques used: immunofluorescence microscopy; confocal microscopy; RT-PCR; Western blotting; molecular cloning; Zebrafish embryo microinjection. References: 1) Bond J, Woods CG. Cytoskeletal genes regulating brain size. Curr Opin Cell Biol. (2006) 18(1):95-101. 2) Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, Costa SM, Baralle D, Raponi M, Karbani G, Rashid Y, Jafri H, Bennett C, Corry P, Walsh CA, Woods CG. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. (2005) 37(4):353-5 3) Lucas EP, Raff JW. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J Cell Biol. (2007) 178(5):725-32. 4) Graser S, Stierhof YD, Nigg EA. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci. (2007) 120:4321-31. 10. Making better treatments for bipolar disorder Dr Robin Williams, School of Biological Sciences, Royal Holloway University London, Egham, Surrey, TW20 0EX. Robin.wiliams@rhul.ac.uk Bipolar disorder (BD) is a devastating disorder giving rise to cyclic changes of mood and often leading to suicide. Understanding the aetiology of the disorder, which is at least partially caused by genetic factors, has so far proved impossible. One of the most well-supported theory describing how bipolar disorder drugs work is called ‘inositol depletion’ (1). This mechanism was first proposed by Berridge et al1 around 20 years ago, and states that the over-activation of a cell signalling pathway in bipolar disorder involves inositol, and that bipolar disorder drugs thus function to deplete cellular inositol. We have published widely on this mechanism, based in a simple biomedical model, Dictyostelium discoideum (2-4), and we successfully applied these and subsequent results to primary mammalian neurons. This project will exploit a cellular response to inositol depletion, whereby cells increase the expression of inositol biosynthetic enzymes. The project will therefore use a promoter for one of these inositol biosynthetic enzymes to express green fluorescent protein. We will then test increased gene expression on exposure to inositol depleting drugs. The outcome from this project will be a mechanism to identify novel compounds with potential efficacy in bipolar disorder treatment, and the isolation of potential new treatments based around the structure of valproic acid. Methods developed will include cell culture, PCR, cloning, cell transformation, DNA isolation, colony screening, fluorescent microscopy, and pharmacology. References: 1) Berridge MJ, Downes CP, Hanley MR. Cell. 1989;59:411-419. 2) Eickholt BJ, Towers G, Ryves WJ et al. Mol Pharmacol. 2005;67:1-8. 3) Shimshoni JA, Dalton EC, Jenkins A et al. Mol Pharmacol. 2007; 71, 884-892. 4) Williams RSB, Cheng L, Mudge AW, Harwood AJ. Nature. 2002;417:292-295.