PRDM9_summary - Przeworski lab
advertisement

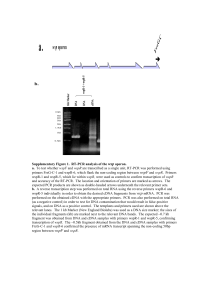
Unpublished PRDM9 sequences for primates Laure Ségurel (lsegurel@mnhn.fr) and Molly Przeworski (mp3284@columbia.edu) Samples: We sequenced 22 PRDM9 alleles from the following individuals: three Western gorillas (Gorilla gorilla), three Sumatran orangutans (Pongo abelii), two siamangs (Symphalangus syndactylus) and three rhesus macaques (Macaca mulatta). The DNA from gorillas, orangutans and siamangs were obtained from the San Diego Zoo and are subject to a Material Transfer Agreement. The macaques DNA were purchased at the Coriell Institute. Data availability: The sequences that we obtained are available on the Przeworski’s lab website (http://przeworski.c2b2.columbia.edu/) Method: The region of PRDM9 containing the zinc finger array was amplified by PCR from 100ng of genomic DNA, using 2U Platinium Hi-Fi Taq DNA polymerase (Invitrogen). The primers used for gorillas, orangutans and siamangs are GTCCCCCGAACACTTACAGA and TGGGCAAGGTTTCAGTGTT. The primers used for macaques are ACTGCTCAGGAGACCAAGGA and TGGGCAAGGTTTCAGTGAT. Cycling conditions were as follows: 94°C for 1 minute, then 30 cycles of 94°C for 30 seconds, 58°C for 1 minute, 68°C for 6 minutes, and a final extension of 68°C for 10 minutes. PCR products were run on 1% TBE agarose gels and the bands corresponding to individual alleles were excised independently, purified using the QIAquick Gel Extraction Kit (Qiagen). They were then cloned into pDONR 221 vector (Invitrogen) and 6 independent inserts were sequenced using the same primers as for PCR. Sequencing was performed using BigDye Terminator v3.1 Cycle Sequencing on an ABI3730 DNA Analyzer (Applied Biosystems) and sequences were analysed using CodonCode Aligner (CodonCode Corporation).