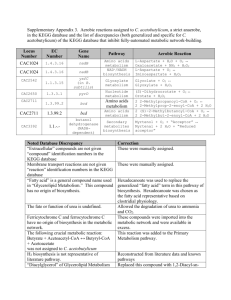
a Plant Molecular Physiology Laboratory, Biology Department
advertisement
Identification and expression analysis of ethylene biosynthesis and signaling genes provide insights on the early and late coffee cultivars ripening pathway Solange A Ságioa, Horllys G Barretoa, André A Limaa, Rafael O Moreira a, Pamela M Rezende a, Luciano V Paivab , Chalfun-Júnior Aa* aPlant Molecular Physiology Laboratory, Biology Department, Federal University of Lavras (UFLA), s/n - Cx. P 3037-37200-000 Minas Gerais, Brazil. bChemistry Department, Central Laboratory of Molecular Biology (LCBM), UFLA, s/n - Cx. P 303737200-000- Minas Gerais, Brazil. *Corresponding author at: Plant Molecular Physiology Laboratory, Biology Department, Federal University of Lavras (UFLA), s/n - Cx. P 3037- Minas Gerais, Brazil. Fax: +55-35-3829-1100 - Email address: chalfunjunior@dbi.ufla.br (A. Chalfun-Jú nior) Suppl. Table 1 Comparison of the coffee ethylene biosynthesis and signaling genes identified in this work with the closest members (clustered in the same clade of the phylogenetic tree (Figure 1)) from the Arabidopsis and tomato ethylene biosynthesis and signaling genes. Coffee sequences marked with an asterisk (*) represent full length cDNAs. The entire sequence from incomplete sequences was used in the aligments Gene Contig/aa ACS CaACS1/231 ACO ETR E-value Identity Similarity 2e-130 167/215(78%) 193/215(89%) SlACS1B (AAF97615) 1e-128 163/215(76%) 192/215(89%) SlACS6 (BAA34923) 1e-127 169/214(79%) 189/214(88%) SlACO1 (P05116) 0.0 271/319(85%) 299/319(93%) SlACO2 (P07920) 0.0 261/320(82%) 289/320(90%) SlACO3 (P24157) 0.0 272/320(85%) 298/320(93%) CaACO4/318* SlACO4 (BAA34924) 0.0 262/318(82%) 290/318(91%) CaACO5/292* SlACO5 (AJ715790.1) 1e-144 194/289(67%) 222/289(76%) AtACO1 (NP179549) 2e-138 185/278(67%) 222/278(79%) AtETR1 (P49333) 0.0 522/617(85%) 569/617(92%) SlETR1 (Q41342) 0.0 530/617(86%) 567/617(91%) CaACO1/319* CaETR1/615 blastp SlACS1A (AAF97614) CaETR3/246 SlETR2 (O49187) 0.0 514/614(84%) 558/614(90%) SlETR3 (Q41341) 1e-100 162/247(66%) 200/247(80%) 8e-95 160/247(65%) 195/247(78%) AtERS1 (Q38846) CTR1 EIN2 EIN3 ERF CaETR4/262 SlETR4 (Q9XET8) 1e-141 201/259(78%) 229/259(88%) CaCTR2/423 SlCTR2 (NP_001234768) 3e-35 55/93(59%) 75/93(80%) SlCTR1 (AAL87456) 5e-25 43/79(54%) 56/79(70%) SlCTR3 (NP_001234454) 2e-26 46/83(55%) 56/83(67%) SlCTR4 (NP_001234457) 1e-20 38/83(46%) 49/83(59%) AtCTR1 (NP_195993) 4e-23 40/69(58%) 50/69(72%) SlEIN2 (AAZ95507) 0.0 288/474(61%) 361/474(76%) AtEIN2 (AAD41076) 3e-128 225/494(46%) 316/494(63%) SlEIL1 (Q94FV4) 0.0 465/634(73%) 531/634(83%) SlEIL4 (Q76DI3) 0.0 462/635(73%) 527/635(82%) SlEIL2 (Q94FV3) 0.0 457/634(72%) 530/634(83%) CaERF1/304* SLERF78 3e-55 109/223(49%) 131/223(58%) CaERF2/304 SLERF79 135/299(45%) 169/299(56%) CaEIN2/483 CaEIN3/644* 7e-65 SLERF80 1e-46 116/263(44%) 142/263(53%) CaERF3/387 SLERF77 2e-117 198/389(51%) 258/389(66%) CaERF4/165 SLERF35 7e-28 54/65(83%) 56/65(86%) CaERF5/404* SLERF77 6e-129 216/405(53%) 273/405(67%) CaERF6/220* SLERF35 5e-59 119/191(62%) 134/191(70%) CaERF7/226 SLERF35 4e-21 49/73(67%) 56/73(76%)