jcpe12432-sup-0001-supinfo
advertisement

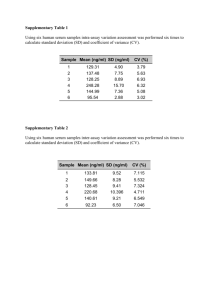
Preliminary insight into the periostin leverage during periodontal tissue healing Miguel Padial-Molina, DDS, PhD, Sarah L. Volk, BS, Hector F. Rios, DDS, PhD Supplementary Methods Clinical measurements The medical and dental histories of the subjects were obtained by a standardized health questionnaire. A total of six sites were measured per tooth (distobuccal, buccal, mesiobuccal, distolingual, lingual, and mesiolingual) at baseline and 4 weeks using a periodontal probe by a calibrated examiner (MP-M). Gingival margin (GM) was measured from the cemento-enamel junction, or another fixed reference point, to the soft tissue margin (positive value for recession; negative value for overgrowth). Pocket depth (PD) was measured as the distance from the gingival margin to the base of the pocket or sulcus. Clinical attachment level (CAL) was calculated as the result of addition of GM and PD. A calibrated UNC probe was used and the distance recorded to the next lowest mm. All teeth present in every arch, except third molars, were measured. Four of the deepest PDs associated with ≥1 mm of CAL and the presence of BOP in each quadrant were selected for sampling in each disease subject. Four random sites were selected in healthy patients. The examiner (MP-M) was compared to the standard examiner on a site basis for PD and CAL in a calibration exercise before the study initiation that included 4 patients (2 disease and 2 healthy). Interand intra-examiner reliability were analyzed. When the difference between the two examiners was within 1mm, the measurements were assumed to be equal. Additional clinical variables recorded include: oral hygiene (OH) (no plaque; plaque detectable with probe; visible plaque; gross accumulation), discomfort (less; less/moderate; moderate; moderate/more; more), soft tissue index (ST) (no inflammation; mild; moderate bleeding; severe bleeding), wound healing index (WHI) (mature; erythema; bleeding; flap mobility; suppuration; necrosis). 1 of 8 Laboratory procedures All samples after collection were immediately assigned a random numerical label so that the laboratory analyses were performed blindly by two experienced laboratory staff (MP-M, SV). PDL and gingival tissue At least three PDL and gingival tissue biopsies were collected from selected sites by an internal bevel gingivectomy performed during open flap procedures in disease sites. The control specimens were collected during gingivectomy or crown lengthening procedures. In both cases, the biopsy consisted of the tissue surrounding the tooth including the gingival margin and the more coronal aspect of the PDL. For ethical reasons, the extent of the biopsy was determined by the extent of the periodontal pocket or the crown lengthening to be performed. Immediately after collection, tissue biopsies were divided into 3 equivalent pieces: 1 piece was immersed in formalin, stored at 4°C overnight and then transferred to 70% ethanol at 4°C until processed for histology; the other 2 pieces were immediately placed in a tube on dry ice and stored at −80°C until assayed for protein and mRNA, respectively. Periostin immunostanining was performed on paraffin embedded sections by using an affinity-purified rabbit polyclonal antibody to periostin (1:8000 dilution in 1% PBS/BSA) (ab14041, Abcam Inc., Cambridge, MA, USA). Immunological reaction was visualized by the use of the UltraVision Quanto Detection System HRP DAB (Thermo Scientific, Fremont, CA, USA) and the sections were counterstained with hematoxylin. Results were obtained by qualitatively assessing immunostaining intensity and distribution. Portions of tissue for mRNA or protein were homogenized by using a tissue homogenizer (Bullet Blender, Next Advance, Inc., Averill Park, NY, USA) in either Trizol or protein extraction buffer containing a protease inhibitor cocktail (Tissue Extraction Reagent, Invitrogen, Life Technologies Corporation, Grand Island, NY, USA). RNA was then isolated (Trizol Plus RNA Purification Kit, Invitrogen). Reverse-transcriptase PCR was performed using a PTC-200 Peltier Thermal Cycler (MJ Research, GMI, Ramsey, MN, USA) immediately after RNA isolation. The RT mix (Taqman Reverse Transcription Reagents kit, Applied Biosystems, Life Technologies) was made for a final volume of 50µl of cDNA (2 µg of RNA and oligo(dT) primer). The probes used in the qPCR (7500 Real Time 2 of 8 PCR System, Applied Biosystems) were POSTN (Hs01566748_m1), ßIG-H3 (Hs00932734_m1), Coll 1 (Hs00164004_m1), Twist (Hs01675818_s1), TGF-ß1 (Hs00998133_m1) and GAPDH (Hs02758991_g1). Each sample was analyzed in triplicate of the cDNA templates. The 2ΔΔCt method was used to calculate gene expression levels relative to GAPDH. Expression of each gene was normalized to the lower periostin expresser. Periostin protein in each sample was analyzed by a sandwich ELISA using recombinant human periostin as a standard (R&D Systems, Inc., Minneapolis, MN, USA), as described by the manufacturer. Results are presented in ng/ml. Whole saliva Unstimulated whole saliva was collected from each subject by the draining method, in which saliva is allowed to drip off the lower lip into a test tube (Navazesh, 1993) until a total of 2ml was collected or 15 minutes had elapsed. The sample was immediately placed on wet ice and transported to the lab. Aprotinin (1mg/ml) at a dilution of 1:100 and phenylmethylsulphonyl fluoride (PMSF) (100 mM in MeOH) at a dilution of 1:200 were added to the sample as protease inhibitors. 400 μl of the prepared WS sample was aliquoted into 5 Eppendorf tubes and stored at −80°C until assayed. Periostin was analyzed in saliva by a sandwich ELISA technique as described above. Gingival crevicular fluid/wound fluid As the selected sites for sampling were subjected to surgery and fluids were collected before and after the surgery and over a period of 4 weeks, we refer to this fluid as GCF/wound fluid. Four different sites per patient were selected as described above for GCF/wound fluid collection. Prior to sampling, the supragingival plaque was removed from the interproximal surfaces with a sterile curette; these surfaces were dried gently with an air syringe and isolated with cotton rolls. The fluid was sampled with filter paper inserted carefully into the crevice until mild resistance was felt and left there for 30 sec. Care was taken to avoid mechanical injury. If resampling was needed at the same site, at least 2 min passed from one to another. Individual samples were placed in Eppendorf tubes and stored at −80°C until assayed. 3 of 8 GCF/wound fluid was extracted from the filter paper by using an extraction solution containing PMSF (1:200 dilution), aprotinin (1:100 dilution) and human serum albumin (1:300 dilution) in 1X PBS. Briefly, 20µl of solution is added onto the white portion of each paper strip that is then secured at the top of a tube using a cap. Tubes are then spun at 2000rpm, 4ºC, for 5 min, a total of 5 times so a final volume of 100µl of sample is obtained. Periostin was analyzed by a sandwich ELISA technique as described above. Results are presented in pg/ml. Additional samples per patient were used to analyze a number of periodontal disease related cytokines (only CRP, IL-1α, IL-1ß, IL-6, IL-8, IL-10, MIP-1α, Osteopontin, Osteoactivin, MMP-9, MMP-13 and TNF-α gave values above the limit of detection) by using an array-based multiplex ELISA system for simultaneous quantitative measurement of multiple cytokines (Human periodontal disease Quantibody Array, RayBiotech, Inc., Norcross, GA, USA). Serum collection Blood sampling was performed by the Michigan Clinical Research Unit. After blood drawn, specimens were clotted for 30 min, centrifuged for 10 min at 2700rpm and serum aliquoted and frozen at -80ºC until assayed. Periostin was detected in diluted serum (1:40 in reagent diluent) by a sandwich ELISA technique as described above. 4 of 8 Supplementary Figures Supplementary Figure S1. Percentages of patients classified by the different categories of categorical clinical variables: A) Oral hygiene; B) Soft tissue index; C) Discomfort; and D) Wound healing index. 5 of 8 Supplementary Figure S2. Levels of periostin protein in tissue in ng/ml (mean±SEM) for healthy (blue) and disease (red) patients. 6 of 8 Supplementary Figure S3. Levels of periostin, ßIG-H3, collagen I, twist and TGF-ß1 mRNA (mean±SEM) in tissue for healthy and disease patients. 7 of 8 Supplementary Figure S4. Levels of CRP over time in serum in mg/dl (mean±SEM) for healthy (blue) and disease (red) patients. 8 of 8