[y 5 ] + - Springer Static Content Server
advertisement
![[y 5 ] + - Springer Static Content Server](http://s3.studylib.net/store/data/007045431_1-1b395830b5d13c2a0484118cf1c75dda-768x994.png)
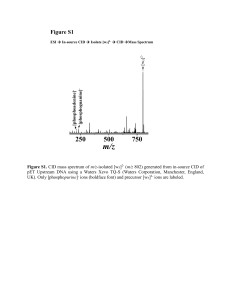
Table S1. Summary of experimental m/z values, mass measurement errors (ppm), and identities of a-“like” product fragment ions generated from CID of m/z-isolated [yn]m+ (n = 6-12; m = 1, 2, and/or 3) species of des-acetylated-α-melanocyte-stimulating hormone and [y5]2+ and [y6]+ species of angiotensin II antipeptide in the LTQ Orbitrap from main text reference 39 (JASMS, 2013, 24, 1755). Isolated y fragment Identity of Fragment Ion Experimental m/z Error (ppm) Des-acetylated-α-melanocyte-stimulating Hormone (SYSMEHFRWGKPV-amidated) 3+ [y12 ] 3+ 12 3+ 12 3+ [y12 - (NH3 + CO)] 497.584 0.26 [y11 ] [y11 - (NH3 + CO)] 443.229 0.09 [y11 ]2+ [y11 - (NH3 + 12 CO)]2+ 664.340 0.42 [y10 ]2+ [y10 - (NH3 + 12 CO)]2+ 620.824 0.45 [y9 ]2+ [y9 - (NH3 + 12 CO)]2+ 555.304 0.14 327.524 0.09 422.253 0.00 348.719 0.17 3+ [y8 ] + [y7 ] 2+ [y6 ] 12 3+ 12 + 12 2+ [y8 - (NH3 + CO)] [y7 - (NH3 + CO)] [y6 - (NH3 + CO)] Angiotensin II Antipeptide (EGVYVHPV) [y7 ]+ [y7 - (H2 O + 12 CO)]+ 724.414 0.52 [y5 ]2+ [y5 - (H2 O + 12 CO)]2+ 284.666 0.18 285.158 0.04 2+ [y5 ] 12 2+ [y5 - (NH3 + CO)] Figure S1 In-source [y5]+ (AAHAA-NH2) Isolation CID Fragments [y5 – (NH3 + 12CO)]+ [y5 – (NH3)]+ [y5 – (NH3 + 13CO)]+ 395 396 m/z 150 [y3]+ 300 [y5 – (NH3 + H2O)]+ [y5 – (NH3 + AA)]+ [y5 – (NH3 + H2O + AA)]+ [y5 – (NH3 + AAA)]+ [y5 – (NH3 + A)]+ [y5 – (NH3 + CO + A)]+ 394 [y5]+ 450 m/z Figure S1. LTQ Orbitrap collision-induced dissociation (CID) mass spectrum of m/z-isolated [y5]+ (m/z 440) (generated from in-source fragmentation of protonated AAAAHAA-NH2). Inset shows the expanded view of the m/z range from 393.5 to 396.5 corresponding to [y5 – (NH3 + CO)]+ (m/z 395) and rearranged product fragment ion [y5 – (NH3 + 13CO)]+ (m/z 394). 12 Figure S2 CID Fragments In-source [y4]+ (AHAA-NH2) Isolation [y4 – (NH3 + 12CO)]+ [y4 – (NH3 + 13CO)]+ 324 325 m/z [y4 – (NH3 + AA)]+ 100 200 300 [y4 – (NH3 + H2O)]+ 323 [y4 – (NH3 + C2NH5)]+ 322 [y4 – (NH3 + A)]+ [y4 – (NH3)]+ 400 m/z Figure S2. LTQ Orbitrap collision-induced dissociation (CID) mass spectrum of m/z-isolated [y4]+ (m/z 369) (generated from in-source fragmentation of protonated AAAAHAA-NH2). Inset shows the expanded view of the m/z range from 322 to 325 corresponding to [y4 – (NH3 + CO)]+ (m/z 324) and rearranged product fragment ion [y4 – (NH3 + 13CO)]+ (m/z 323). 12 Figure S3 ESI [M + H]+ (AAAAHAA-NH2) Isolation (a) [a5]+ or [b5 – (12CO)]+ [a5]+ or [b5 – (13CO)]+ CID Fragments [a6]+ or [b6 – (12CO)]+ (b) [a6]+ or [b6 – (13CO)]+ [b7]+ 395 396 m/z (c) 464 465 466 467 m/z [a7]+ or [b7 – (12CO)]+ [a7]+ or [b7 – (13CO)]+ [b6]+ 536 [b5]+ 537 538 [y3]+ [b4]+[y4]+ (a) (b) (c) [b7 – (H2O)]+ 394 [b6 – (H2O)]+ 393 m/z 200 400 600 m/z Figure S3. LTQ Orbitrap collision-induced dissociation (CID) mass spectrum of m/z-isolated protonated molecular ion of AAAAHAA-NH2 (m/z 582). Insets show the expanded views of the m/z ranges from (a) 393 to 396 corresponding to [a5]+ ([b5 – (12CO)]+) (m/z 395) and rearranged product fragment ion [a5]+ ([b5 – (13CO)]+) (m/z 394), (b) 464 to 467 corresponding to [a6]+ ([b6 – (12CO)]+) (m/z 466) and rearranged product fragment ion [a6]+ ([b6 – (13CO)]+) (m/z 465), and (c) 535.5 to 538.5 corresponding to [a7]+ ([b7 – (12CO)]+) (m/z 537) and rearranged product fragment ion [a7]+ ([b7 – (13CO)]+) (m/z 536). Please note the change in ratios of 13CO to 12CO losses as a function of fragment ion size (insets). In smaller a-type ions, there is a higher probability of losing the 13 CO (or the entire labeled alanine (i.e., A)) because fewer unlabeled backbone carbonyl groups are available. Because the precursor peptide contains five isobaric alanines, there are (at least) two possible pathways for generation of the a-type ions in Figure S3. Pathway I: Isolation of [M + H]+ (m/z 582) CID Generation of Normal a-Type Ions. Pathway II: Isolation of [M + H]+ (m/z 582) CID Generation of y-Type Ions Gas-phase Rearrangement Generation of a-“Like” Ions via main text Scheme 1 and/or 2. Results from CID of m/z-isolated y-type ions (Figure 1, S1, and S2) suggest that pathway II may contribute to the CID mass spectrum of protonated molecular ions. Therefore, sequence-scrambling through gas-phase dissociation of y-type ions should not be negated. Figure S4 In-source [y7]+ (EGVYVHPV) Isolation Pre-IM IM CID IM Spectrum 4.0 4.5 5.0 5.5 6.0 6.5 Arrival Time (ms) Figure S4. Synapt G2-S HDMS ion mobility (IM) profile showing multiple structural isomers for [y7 – (H2O + 12 CO)]+ (m/z 724) generated from pre-IM CID of [y7]+ (m/z 770) from angiotensin II antipeptide (amino acid sequence: EGVYVHPV). Product fragment ions from [y7]+ of angiotensin II antipeptide were generated prior to IM separation (pre-IM CID) in the trap cell using collision energies corresponding to a 38 V potential difference between the trap cell exit and entrance of the helium cell. The potential difference between the exit of the IM cell and entrance of the transfer cell was set at 2.0 V to prevent post-IM fragmentation of ions of interest. IM profile corresponds to the average of 2400 scans. All other instrument parameters were kept similar to those used for IM-MS analysis of des-acetylated-α-melanocyte-stimulating hormone (see main text for details). Figure S5 In-source [y5]+ (DRVYIHPF) Isolation Pre-IM IM CID IM Spectrum 3.0 3.5 4.0 4.5 5.0 5.5 Arrival Time (ms) Figure S5. Synapt G2-S HDMS ion mobility (IM) profile showing multiple structural isomers for [y5 – (H2O + 12 CO)]+ (m/z 630) generated from pre-IM CID of [y5]+ (m/z 676) from angiotensin II, human (amino acid sequence: DRVYIHPF). Product fragment ions from [y5]+ of angiotensin II were generated prior to IM separation (pre-IM CID) in the trap cell using collision energies corresponding to a 30 V potential difference between the trap cell exit and entrance of the helium cell. The potential difference between the exit of the IM cell and entrance of the transfer cell was set at 2.0 V to prevent post-IM fragmentation of ions of interest. IM profile corresponds to the average of 1200 scans. Source offset was set to 85 V. Nitrogen (N2) gas pressure in the IM cell was ~4.0 mbar. IM wave velocity was set to 500 m/s. All other instrument parameters were kept similar to those used for IM-MS analysis of des-acetylated-α-melanocytestimulating hormone (see main text for details).