tpj12135-sup-0001-FigureS1-S7
advertisement

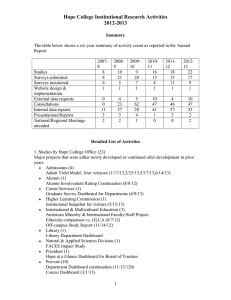
Figure S1. Phylogenetic analysis of the GT8 GUX clade proteins. Bootstrapped phylogeny based on the putative catalytic domain of identified AtGUX1-5 orthologues. Lower plants are represented by S. moellendorffii (red) and P. patens (orange). Monocots are represented by O. sativa, S. bicolor, B. distachyon and Z. mays (shades of green), while dicots are represented by E. grandis, P. trichocarpa and A. thaliana (shades of blue). Asterisks mark Arabidopsis proteins studied in this report. Figure S2. DASH analyses used to quantify fragments released by XynC hydrolysis of xylan from Col0, gux1 and gux2. XynC-released oligosaccharides were derivatised with APTS and separated by capillary electrophoresis (DASH). Shorter UXn oligosaccharides could be resolved into GlcA and MeGlcAdecorated oligosaccharides (inset), and these were combined for quantitation. Figure S3. MALDI-ToF MS spectra of XynC hydrolysis products. MALDI-TOF-MS spectra showing signals ([M+Na]+, [M-H+2Na]+ and [M-2H+3Na]+) for the 2-AA labelled acidic oligosaccharides produced from digestion of Arabidopsis xylan with XynC. Doubly and trebly sodiated oligosaccharides are labelled with coloured circles (black and red- doubly sodiated oligosaccharides with GlcA or MeGlcA respectively; blue and yellow- trebly sodiated oligosaccharides with GlcA or MeGlcA respectively). Figure S4. Structural characterization of a XynC oligosaccharide with high energy MALDI-CID. XynC-released oligosaccharides were reductively aminated with 2-AA, separated by HILIC and structurally characterised using MALDI-TOF-MS with collision induced dissociation (CID). The fragmentation spectrum of 2-AA labelled UX6 is shown. The series of Y, B and 1,5X ions indicate the presence of a branching (which corresponds to a MeGlcA) on the second Xylp from the reducing end. This is confirmed by the presence of the non-reducing 3,5 A ions, which also indicate the presence of (1→4) linkages between the Xylp residues. Glycosidic and cross-ring fragments are identified according to Domon and Costello nomenclature (Domon and Costello, 1988). The ‘elimination’ ion incorporating the reducing end, defined here as V5α (see inset for the proposed chemical structure for this ‘elimination’ ion), shows the presence of the methyl modification at the C-4 position of GlcA. Two other important ‘elimination’ ions (Spina et al., 2004) G2 (m/z 582.3) and E5 (m/z 667.3) indicate that the 4-O-Me-GlcA is linked at the C-2 position of Xylp. The presence of the H5 (m/z 699.3) sugar lactone (Maslen et al., 2007) confirms this assignment. Hence the oligosaccharide was identified as β-D-Xylp-(1→4)-β-D-Xylp-(1→4)-β-D-Xylp-(1→4)-β-D-Xylp-(1→4)-[α-D-4-O-Me-GlcA-(1→2)]-β-DXylp-(1→4)-D-Xylp. Figure S5. Analysis of [Me]GlcA spacing in xylan from WT and wat1 mutant stems. wat1 mutant plants were grown under short day conditions and therefore lacked fibre cells. Xylan in AIR was hydrolysed with XynC analysed and quantitated by DASH to investigate [Me]GlcA spacing in xylem vessels. Values presented are means ± SD, and are from three XynC hydrolyses of two biological replicates. Figure S6. Expression of GUX1-myc and GUX2-myc in Arabidopsis WT and mutants. Proteins in inflorescence stems of WT, gux1, gux2 and gux 1 gux2 double mutant and plants transformed to express pIRX9:GUX1 or pIRX9:GUX2 were extracted and then separated by SDS-PAGE and myc-tagged proteins detected by Western Blot. The predicted molecular mass of GUX1-myc is 85 kDa, and of GUX2-myc is 79 kDa. Figure S7. Monosaccharide analysis of Arabidopsis lines over-expressing GUX1 and GUX2 AIR from transformed lines and their backgrounds were hydrolysed to constituent monosaccharides with 2M TFA and analysed by HPAEC-PAD. (Inset) Adjusted scale for GlcA. Domon B, Costello CE (1988) A systematic nomenclature for carbohydrate fragmentations in FABMS/MS spectra of glycoconjugates. Glycoconjugate Journal 5: 397-409 Maslen SL, Goubet F, Adam A, Dupree P, Stephens E (2007) Structure elucidation of arabinoxylan isomers by normal phase HPLC-MALDI-TOF/TOF-MS/MS. Carbohydrate Research 342: 724735 Spina E, Sturiale L, Romeo D, Impallomeni G, Garozzo D, Waidelich D, Glueckmann M (2004) New fragmentation mechanisms in matrix-assisted laser desorption/ionization time-offlight/time-of-flight tandem mass spectrometry of carbohydrates. Rapid Communications in Mass Spectrometry 18: 392-398