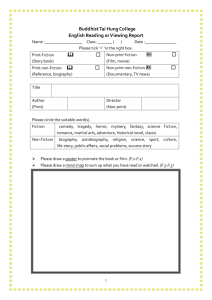
Cells have evolved nucleotide modifications that alter the chemistry
advertisement

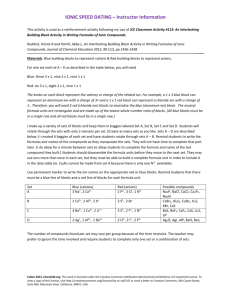
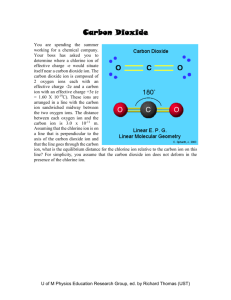
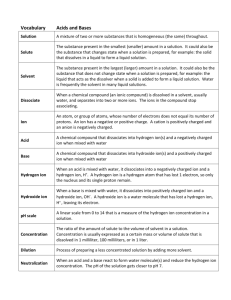
Charge Reversal Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Vladislav V. Lobodin1, Joshua J. Savory1, Nathan K. Kaiser1, Paul W. Dunk2, and Alan G. Marshall1,2 1 2 National High Magnetic Field Laboratory, 1800 East Paul Dirac Drive, Tallahassee, FL 32310, USA Florida State University, Department of Chemistry and Biochemistry, 95 Chieftain Way, Tallahassee, FL 32306, USA Supporting Information 1 Table S1. Elemental composition assignments for charge reversal ions from 3,3’bicarbazole. m/z 333.1297 332.1263 332.1218 331.1230 331.1186 331.1142 330.1153 330.1108 329.1074 329.1029 328.0996 328.0951 327.0918 327.0873 326.0839 326.0795 325.0761 324.0682 323.0605 316.0994 315.0916 314.0964 314.0838 305.1073 304.1121 304.0994 304.0950 303.1042 303.0998 303.0917 302.0964 302.0921 302.0839 301.0886 301.0842 301.0761 300.0888 300.0808 300.0764 300.0683 299.0730 Rel. Magnitude (%) 3.5 28.1 1.6 100.0 13.5 1.3 57.4 16.7 67.3 6.3 26.4 6.5 26.0 3.0 11.9 1.4 5.3 3.3 1.1 1.7 1.7 1.0 1.0 1.9 1.7 1.3 1.1 3.6 2.1 4.6 7.8 3.7 3.3 14.2 1.4 2.4 1.1 5.4 2.5 1.8 10.3 Elemental Composition [13C2C22H15N2]+ [13CC23H15N2]+ [13C2C22H14N2]+ [C24H15N2]+ [13CC23H14N2]+ [13C2C22H13N2]+ [C24H14N2]+ [13CC23H13N2]+ [C24H13N2]+ [13CC23H12N2]+ [C24H12N2]+ [13CC23H11N2]+ [C24H11N2]+ [13CC23H10N2]+ [C24H10N2]+ [13CC23H9N2]+ [C24H9N2]+ [C24H8N2]+ [C24H7N2]+ [C23H12N2]+ [C23H11N2]+ [C24H12N]+ [C23H10N2]+ [C22H13N2]+ [C23H14N]+ [C22H12N2]+ [13CC21H11N2]+ [C23H13N]+ [13CC22H12N]+ [C22H11N2]+ [C23H12N]+ [13CC22H11N]+ [C22H10N2]+ [C23H11N]+ [13CC22H10N]+ [C22H9N2]+ [13CC23H11]+ [C23H10N]+ [13CC22H9N]+ [C22H8N2]+ [C23H9N]+ 2 Mass Error, (ppm) 0.2 0.0 -0.1 0.0 0.3 0.4 0.5 0.3 0.2 0.2 0.2 0.3 0.4 0.3 0.3 0.3 0.1 0.1 0.3 -0.3 -0.1 -0.1 -0.3 -0.1 -0.1 -0.3 -0.1 -0.3 0.0 -0.1 0.0 0.3 0.1 0.0 0.2 0.3 -0.2 0.1 0.3 0.2 0.2 299.0685 298.0652 297.0574 296.0495 288.0808 278.0964 277.0886 276.0807 275.0730 274.0777 274.0651 273.0573 272.0621 270.0463 1.5 3.4 3.7 1.3 1.4 1.8 2.7 1.6 3.9 2.8 1.6 2.4 2.2 1.3 [13CC22H8N]+ [C23H8N]+ [C23H7N]+ [C23H6N]+ [C22H10N]+ [C21H12N]+ [C21H11N]+ [C21H10N]+ [C21H9N]+ [C22H10]+ [C21H8N]+ [C21H7N]+ [C22H8]+ [C22H6]+ 3 0.1 0.2 0.3 0.1 0.0 -0.2 -0.2 -0.3 0.0 0.0 0.1 0.0 0.0 -0.4 Figure S1. Schematic diagram of a 7-section open cylindrical cell. Dimensions of segments are in millimeters. The angular extent of each detection and excitation electrode is 120º and 60 º for optimal detection. Figure S2. The instrument set-up for charge reversal experiments induced by electron ionization (top) and UV irradiation (bottom). Figure S3. Potential energy profiles for a negative ion (bottom) and the positive ion (top) formed by essentially vertical (i.e., Franck-Condon) excitation. Because the positive product ion is formed in an excited state, it may readily fragment (see text). Figure S4. Trajectory of a cyclotron-excited ion before (blue) and immediately after (red) charge reversal, as a result of collision with a gas molecule. The post-excitation radius determines precursor ion Ekin and its Eint after collision (see text). Moreover, the precursor negative ion maximum post-excitation radius is clearly 1/3 of the cell radius. 4