Additional file 8
advertisement
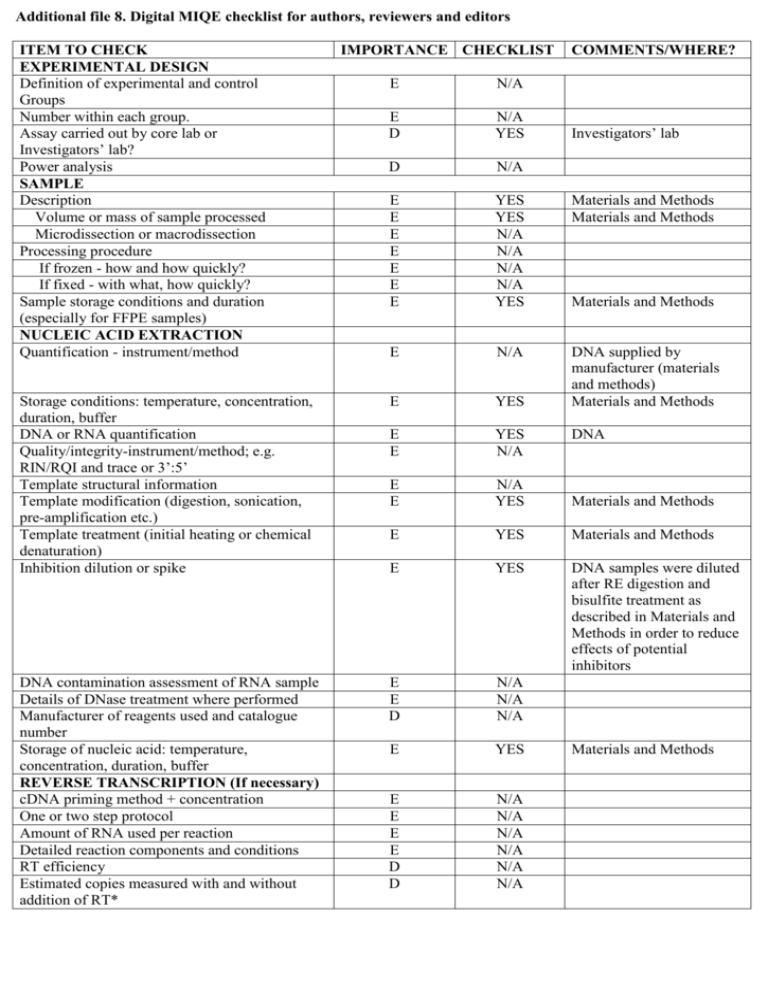
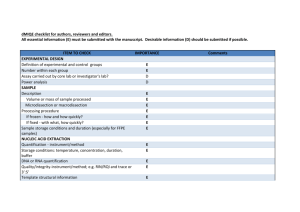
Additional file 8. Digital MIQE checklist for authors, reviewers and editors ITEM TO CHECK EXPERIMENTAL DESIGN Definition of experimental and control Groups Number within each group. Assay carried out by core lab or Investigators’ lab? Power analysis SAMPLE Description Volume or mass of sample processed Microdissection or macrodissection Processing procedure If frozen - how and how quickly? If fixed - with what, how quickly? Sample storage conditions and duration (especially for FFPE samples) NUCLEIC ACID EXTRACTION Quantification - instrument/method IMPORTANCE CHECKLIST COMMENTS/WHERE? E N/A E D N/A YES D N/A E E E E E E E YES YES N/A N/A N/A N/A YES Materials and Methods Materials and Methods E N/A Storage conditions: temperature, concentration, duration, buffer DNA or RNA quantification Quality/integrity-instrument/method; e.g. RIN/RQI and trace or 3’:5’ Template structural information Template modification (digestion, sonication, pre-amplification etc.) Template treatment (initial heating or chemical denaturation) Inhibition dilution or spike E YES DNA supplied by manufacturer (materials and methods) Materials and Methods E E YES N/A E E N/A YES Materials and Methods E YES Materials and Methods E YES DNA samples were diluted after RE digestion and bisulfite treatment as described in Materials and Methods in order to reduce effects of potential inhibitors DNA contamination assessment of RNA sample Details of DNase treatment where performed Manufacturer of reagents used and catalogue number Storage of nucleic acid: temperature, concentration, duration, buffer REVERSE TRANSCRIPTION (If necessary) cDNA priming method + concentration One or two step protocol Amount of RNA used per reaction Detailed reaction components and conditions RT efficiency Estimated copies measured with and without addition of RT* E E D N/A N/A N/A E YES E E E E D D N/A N/A N/A N/A N/A N/A Investigators’ lab Materials and Methods DNA Materials and Methods Manufacturer of reagents used and catalogue number Reaction volume (for two step reverse transcription reaction) Storage of cDNA: temperature, concentration, duration, buffer dPCR TARGET INFORMATION Sequence accession number D N/A D N/A D N/A E YES Location of amplicon Amplicon length In silico specificity screen (BLAST, etc) Pseudogenes, retropseudogenes or other homologs? Sequence alignment Secondary structure analysis of amplicon and GC content Location of each primer by exon or intron (if applicable) Where appropriate, which splice variants are targeted? dPCR OLIGONUCLEOTIDES Primer sequences and/or amplicon context sequence** RTPrimerDB Identification Number Probe sequences** Location and identity of any modifications Manufacturer of oligonucleotides Purification method dPCR PROTOCOL Complete reaction conditions Reaction volume and amount of RNA/cDNA/DNA Primer, (probe), Mg++ and dNTP concentrations Polymerase identity and concentration D E E D YES YES YES YES D D YES N/A Available on request E YES Additional file 6 E N/A E YES Additional file 6 D D E D D N/A YES YES YES YES Additional file 6 Additional file 6 Materials and Methods HPLC E E YES YES Materials and Methods Materials and Methods E YES Materials and Methods; Manufacturer’s proprietary E YES AmpliTaq Gold® DNA Polymerase; concentration is Manufacturer’s proprietary Buffer/kit Catalogue No and manufacturer Exact chemical constitution of the buffer Additives (SYBR Green I, DMSO, etc.) Plates/tubes Catalogue No and manufacturer Complete thermocycling parameters Reaction setup Gravimetric or volumetric dilutions (manual/robotic) Total PCR reaction volume prepared Partition number Individual partition volume Total volume of the partitions measured (effective reaction size) E D E D E D D YES NO N/A YES YES YES YES Materials and Methods Manufacturers’ proprietary D E E E YES YES YES YES Materials and Methods Materials and Methods 0.85 nL 770 partitions (digital Methylight) 3080 partitions (MDRE/MSRE dPCR) CDKN2A (p14ARF): NC_000009.12 COL2A1: NC_000012.12 Additional file 6 Additional file 6 Available on request None detected by BLASTn Materials and Methods Materials and Methods Materials and Methods Manual Partition volume variance/standard deviation Comprehensive details and appropriate use of controls Manufacturer of dPCR instrument dPCR VALIDATION Optimisation data for the assay Specificity (when measuring rare mutations, pathogen sequences etc.) Limit of detection of calibration control If multiplexing, comparison with singleplex assays DATA ANALYSIS Average copies per partition (λ or equivalent ) dPCR analysis program (source, version) Outlier identification and disposition Results of NTCs D E N/A YES E YES D E YES N/A D E N/A YES E E E E YES YES N/A YES Examples of positive(s) and negative experimental results as supplemental data Where appropriate, justification of number and choice of reference genes E YES E YES Where appropriate, description of normalisation method Number and concordance of biological replicates Number and stage (RT or qPCR) of technical replicates Repeatability (intra-assay variation) E YES COL2A1 assay has been previously described as an effective methylation independent reference control gene for Methylight [40,56,57]. Materials and Methods D E N/A YES Materials and Methods E N/A See below reproducibility analysis Reproducibility (inter-assay/user/lab etc. variation ) Experimental variance or confidence interval*** Statistical methods used for analysis D YES E E YES YES Standard deviation measurements in Results Results and Additional file 4 Data submission using RDML D N/A Materials and Methods and Results Materials and Methods Available on request Results and Discussion, Additional file 9 Results, Additional file 4 Materials and Methods All NTCs gave negative results as shown in Additional file 9 Additional file 9 Materials and Methods and Results All essential information (E) must be submitted with the manuscript. Desirable information (D) should be submitted if possible. * Assessing the absence of DNA using a no RT assay (or where RT has been inactivated) is essential when first extracting RNA. Once the sample has been validated as DNA-free, inclusion of a no-RT control is desirable, but no longer essential. ** Disclosure of the primer and probe sequence is highly desirable and strongly encouraged. However, since not all commercial pre-designed assay vendors provide this information when it is not available assay context sequences must be submitted [58]. *** When single dPCR experiments are performed, the variation due to counting error alone should be calculated from the binomial (or suitable equivalent) distribution.