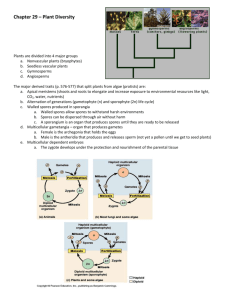
here - WordPress.com

#### Supplement 4: R code for simulating the effect of chytridiomycosis on survival rates ####
### Settings wtemp_kc = c(13.94736842, 14.87037037, 16.61290323, 19.93333333, 16.6, 14.81481481) # Mean water temperature (C) at midnight for each month of the active season at the Kalkallo Creek, Donnybrook wtemp_mcd = c(15.26470588, 17.76666667, 18.87096774, 18.96774194, 18.83928571, 16.0483871) # Mean water temperature (C) at midnight for each month of the active season at the Merri Creek, Donnybrook wtemp_brq = c(19.11111111, 19.51666667, 22.62903226, 23.79032258, 19.55357143, 20.32142857) # Mean water temperature (C) at midnight for each month of the active season at the Campbellfield quarry age = 1 # Frog is an sub-adult or adult flood = 0 # No flood month.days <- c(31,30,31,31,28,31) # Number of days in each month of the active season
### Parameters
## Infection probability site.eff_kc = rnorm(10000, -1.91, 0.2665) # Site-level random effect for Kalkallo Creek site.eff_mcd = rnorm(10000, -1.875, 0.3382) # Site-level random effect for Merri Creek site.eff_brq = rnorm(10000, -2.431, 0.4268) # Site-level random effect for Campbellfield quarry
beta.wtemp = rnorm(10000, -1.082, 0.2189) # Effect of water temperature on infection probability beta.age = rnorm(10000, 1.364, 0.2917) # Effect of frog age on infection probability
## Zoospore load site.eff.spores_kc = rnorm(10000, 4.956, 0.5859) # Site-level random effect for Kalkallo Creek site.eff.spores_mcd = rnorm(10000, 4.358, 0.6463) # Site-level random effect for Merri Creek site.eff.spores_brq = rnorm(10000, 3.731, 0.8979) # Site-level random effect for Campbellfield quarry beta.age.spores = rnorm(10000, 0.3585, 0.6457) # Effect of frog age on zoospore load beta.wtemp.spores = rnorm(10000, -1.481, 0.4392) # Effect of water temperature on zoospore load sigma.resid = rnorm(10000, 2.447, 0.1496) # Residual variation
## Survival probability alpha.surv = rnorm(10000, 4.629, 0.1555) # Intercept for the probability of infection beta.chy = rnorm(10000, -0.478, 0.2071) # Effect of zoospore load on the probability of survival beta.flood = rnorm(10000, -0.618, 0.4664) # Effect of flood on the probability of survival
## Number of parameter combinations ncombinations = length(site.eff_kc)
## Number of simulations nsims = 1000
### Example simulation - October at Kalkallo Creek, Donnybrook
## With chytrid
# Function
FUNC = function(site.eff_kc, beta.wtemp, beta.age, beta.age.spores, beta.wtemp.spores, site.eff.spores_kc, sigma.resid,
alpha.surv, beta.chy, beta.flood){
logit.inf = p.inf = inf.stat = rep(NA, nsims) # Set up vectors
spores = n.spores = n.spores.inf = rep(NA, nsims)
surv = logit.surv = p.surv = p.month.surv = rep(NA, nsims)
for(i in 1:nsims){
logit.inf[i] = site.eff_kc + beta.age * age + beta.wtemp * ((wtemp_kc[1] - 21.68) / (2 * 2.761)) # Infection probability for each frog determined by site-level random effect, frog age and water temperature at that site in that month
p.inf[i] = sapply(logit.inf[i], plogis) # Convert to probability scale
inf.stat[i] = rbinom(1, 1, p.inf[i]) # Infection status a Bernoulli variable
spores[i] = site.eff.spores_kc + beta.age.spores * age + beta.wtemp.spores * ((wtemp_kc[1] - 20.63) / (2 * 3.097)) # Zoospore load for frog determined by site-level random effect, frog age and water temperature at that site in that month
n.spores[i] = rnorm(1, spores[i], sigma.resid) # Zoospore load drawn from a normal distribution
n.spores.inf[i] = inf.stat[i] * n.spores[i] # Only infected individuals have a zoospore load
logit.surv[i] = alpha.surv + beta.chy * ((n.spores.inf[i] - 1.385) / (2 * 2.602)) + beta.flood * flood # Daily probability of survival for each frog is a function of infection status and load, plus flood occurrence
p.surv[i] = sapply(logit.surv[i], plogis) # Convert to probability scale
p.month.surv[i] = p.surv[i] ^ month.days[1] # Calculate probability of survival across the month, by raising the daily probability of survival to the power of number of days in the month
surv[i] = rbinom(1, 1, p.month.surv[i]) # Survival or not is a Bernoulli variable
}
prsurv = sum(surv)/nsims # Probability of survival is the proportion of simulations in which survival occurs
return(prsurv)
}
# Run simulation results <- sapply(1:ncombinations, function(x) {
message('Parameter combination ', x, ' of ', ncombinations, '\n') # Progress indicator
replicate(1, FUNC(site.eff_kc[x], beta.wtemp[x], beta.age[x], beta.age.spores[x], beta.wtemp.spores[x], site.eff.spores_kc[x], sigma.resid[x],
alpha.surv[x], beta.chy[x], beta.flood[x])) # Run FUNC for each parameter combination (x).
}) surv.chytrid.oct_kc = results
## Without chytrid
# Function (exactly the same, for simplicity, but with the probability of infection set to zero)
FUNC = function(site.eff_kc, beta.wtemp, beta.age, beta.age.spores, beta.wtemp.spores, site.eff.spores_kc, sigma.resid,
alpha.surv, beta.chy, beta.flood){
logit.inf = p.inf = inf.stat = rep(NA, nsims) # Set up vectors
spores = n.spores = n.spores.inf = rep(NA, nsims)
surv = logit.surv = p.surv = p.month.surv = rep(NA, nsims)
for(i in 1:nsims){
logit.inf[i] = site.eff_kc + beta.age * age + beta.wtemp * ((wtemp_kc[1] - 21.68) / (2 * 2.761)) # Infection probability for each frog determined by site-level random effect, frog age and water temperature at that site in that month
p.inf[i] = sapply(logit.inf[i], plogis) * 0 # Convert to probability scale, and set to zero to ensure no infections
inf.stat[i] = rbinom(1, 1, p.inf[i]) # Infection status a Bernoulli variable
spores[i] = site.eff.spores_kc + beta.age.spores * age + beta.wtemp.spores * ((wtemp_kc[1] - 20.63) / (2 * 3.097)) # Zoospore load for frog determined by site-level random effect, frog age and water temperature at that site in that month
n.spores[i] = rnorm(1, spores[i], sigma.resid) # Zoospore load drawn from a normal distribution
n.spores.inf[i] = inf.stat[i] * n.spores[i] # Only infected individuals have a zoospore load
logit.surv[i] = alpha.surv + beta.chy * ((n.spores.inf[i] - 1.385) / (2 * 2.602)) + beta.flood * flood # Daily probability of survival for each frog is a function of infection status and load, plus flood occurrence
p.surv[i] = sapply(logit.surv[i], plogis) # Convert to probability scale
p.month.surv[i] = p.surv[i] ^ month.days[1] # Calculate probability of survival across the month, by raising the daily probability of survival to the power of number of days in the month
surv[i] = rbinom(1, 1, p.month.surv[i]) # Survival or not is a Bernoulli variable
}
}
prsurv = sum(surv)/nsims # Probability of survival is the proportion of simulations in which survival occurs
return(prsurv)
# Run simulation results <- sapply(1:ncombinations, function(x) {
message('Parameter combination ', x, ' of ', ncombinations, '\n') # Progress indicator
replicate(1, FUNC(site.eff_kc[x], beta.wtemp[x], beta.age[x], beta.age.spores[x], beta.wtemp.spores[x], site.eff.spores_kc[x], sigma.resid[x],
alpha.surv[x], beta.chy[x], beta.flood[x])) # Run FUNC for each parameter combination (x).
}) surv.chytrid.free.oct_kc = results
## Calculate difference in probability of survival between chytrid and chytrid-free scenarios, for each parameter combination delta.surv.oct_kc = surv.chytrid.oct_kc - surv.chytrid.free.oct_kc
## Calculate summary mean = mean(delta.surv.oct_kc) lo = quantile(delta.surv.oct_kc, 0.025) up = quantile(delta.surv.oct_kc, 0.975) summary.oct_kc = cbind(mean, lo, up) print(summary.oct_kc)