Natronorubrum sulfidifaciens sp
advertisement
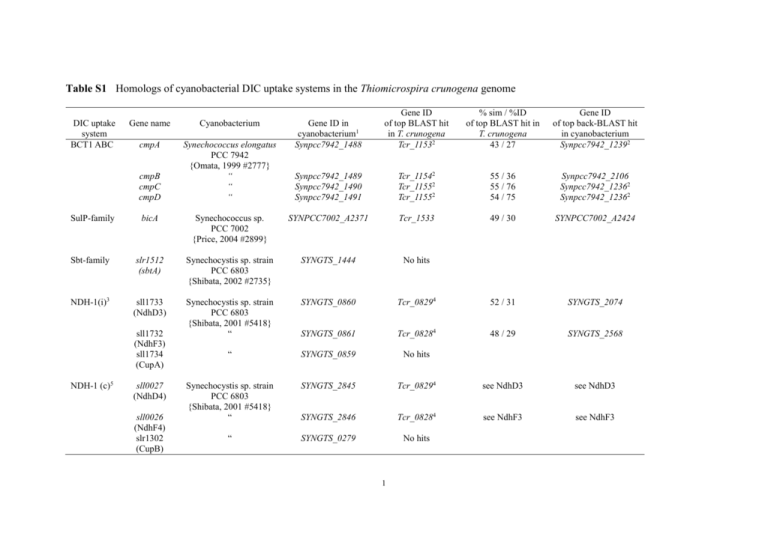
Table S1 Homologs of cyanobacterial DIC uptake systems in the Thiomicrospira crunogena genome
DIC uptake
system
BCT1 ABC
Gene name
Cyanobacterium
cmpA
Synechococcus elongatus
PCC 7942
{Omata, 1999 #2777}
“
“
“
cmpB
cmpC
cmpD
Gene ID in
cyanobacterium1
Synpcc7942_1488
Gene ID
of top BLAST hit
in T. crunogena
Tcr_11532
% sim / %ID
of top BLAST hit in
T. crunogena
43 / 27
Gene ID
of top back-BLAST hit
in cyanobacterium
Synpcc7942_12392
Synpcc7942_1489
Synpcc7942_1490
Synpcc7942_1491
Tcr_11542
Tcr_11552
Tcr_11552
55 / 36
55 / 76
54 / 75
Synpcc7942_2106
Synpcc7942_12362
Synpcc7942_12362
49 / 30
SYNPCC7002_A2424
SulP-family
bicA
Synechococcus sp.
PCC 7002
{Price, 2004 #2899}
SYNPCC7002_A2371
Tcr_1533
Sbt-family
slr1512
(sbtA)
Synechocystis sp. strain
PCC 6803
{Shibata, 2002 #2735}
SYNGTS_1444
No hits
NDH-1(i)3
sll1733
(NdhD3)
Synechocystis sp. strain
PCC 6803
{Shibata, 2001 #5418}
“
SYNGTS_0860
Tcr_08294
52 / 31
SYNGTS_2074
SYNGTS_0861
Tcr_08284
48 / 29
SYNGTS_2568
“
SYNGTS_0859
No hits
Synechocystis sp. strain
PCC 6803
{Shibata, 2001 #5418}
“
SYNGTS_2845
Tcr_08294
see NdhD3
see NdhD3
SYNGTS_2846
Tcr_08284
see NdhF3
see NdhF3
“
SYNGTS_0279
No hits
sll1732
(NdhF3)
sll1734
(CupA)
NDH-1 (c)5
sll0027
(NdhD4)
sll0026
(NdhF4)
slr1302
(CupB)
1
1
Locus tags are from IMG (http://img.jgi.doe.gov)
Part of an apparent operon with genes associated with assimilatory nitrate reduction (e.g., nitrite reductase; nitrate reductase)
3
Inducible by growth under low CO2 conditions
4
Part of an apparent operon including all the other genes of the NDH-1 complex, and may be part of the respiratory NDH-1 complex
5
Constitutively expressed
2
2
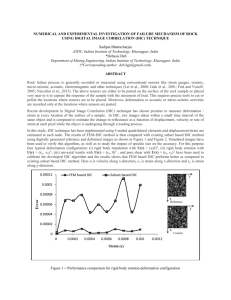
Fig. S1. Response of carbon fixation rate to the amount of purified RubisCO added to
the assay. (A) Timecourse of CO2 fixation by incubations with different RubisCO
concentrations; (B) Carbon fixation rates calculated from the timecourses in (A).
Error bars indicate the standard error of the CO2 fixation rates.
3
2.5
Fixed Carbon
Intracellular DIC
(mM)
2
1.5
1
0.5
0
-TS
+TS
+CCCP
+DCCD
-TS
+TS
+CCCP
+DCCD
3
(mM)
2.5
2
1.5
1
0.5
0
8
(mM)
6
4
2
0
-TS
+TS
+CCCP
+DCCD
Fig. S2. Effects of thiosulfate and metabolic inhibitors on the concentrations of
intracellular fixed carbon and dissolved inorganic carbon (DIC) in Thiomicrospira
crunogena cells incubated in the presence of 0.1 mM DIC (3 individual cultures). ‘TS’ = 0 mM thiosulfate, ‘+TS’ = 40 mM thiosulfate, ‘+CCCP’ = 40 mM thiosulfate +
10 M carbonyl-cyanide m-chlorophenyl hydrazine, ‘+DCCD’ = 40 mM thiosulfate +
1 mM N,N’-dicyclohexylcarbodiimine. Data from cells grown independently in three
chemostats are shown. Error bars indicate standard errors.
4
Fig. S3. Models for DIC uptake consistent with dependence on ATP and proton
potential. A) DIC uptake is mediated by a single transporter that directly couples
DIC uptake to ATP hydrolysis. B) A secondary DIC transporter (relying on proton
potential and/or to drive DIC uptake) works in tandem with an ATP-sensitive DIC
transporter. C) Tandem activities of a cation ATPase and a cation-DIC symporter are
responsible for DIC uptake. D) A C4-like pathway could operate {Hatch, 1987
#2094}, in which an ATP-dependent carboxylase ‘captures’ DIC, which is
subsequently released near or within the carboxysome (c’some) by a decarboxylase.
5
Fig. S4. Thiomicrospira crunogena homologs of genes encoding the cyanobacterial
BCT1 ABC bicarbonate transporter. (A) Gene cluster encoding BCT1 homologs in
6
the T. crunogena genome (http://img.jgi.doe.gov). The gene shaded grey encodes
the homolog of the periplasmic solute-binding component of BCT1 (cmpA) and
corresponds to IMG Gene ID # 637785877. (B) Maximum likelihood analysis of the
cmpA homolog from T. crunogena (marked with an arrow). Amino acid sequences
retrieved from IMG were aligned using MUSCLE and GBLOCKS and phylogenetic
analysis was implemented in MEGA 5. Genes whose products have been
biochemically characterized with respect to substrate specificity are marked with
asterisks. ‘Nitred’ in the taxon title indicates that the gene is present in a likely operon
with genes encoding nitrate reductase. Bootstrap values were computed from from
1000 resamplings of the alignment.
7