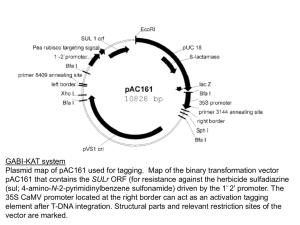
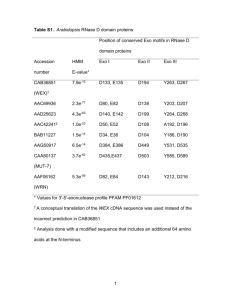
tpj12182-sup-0003-SupportingLegends
advertisement

Supporting information Figure S1. Molecular characterisation of the rad51d mutant. The SAIL_564_A06 line carrying a T-DNA insertion in the Arabidopsis RAD51D coding sequence was found by interrogating the SIGnAL T-DNA express database (Alonso et al., 2003). To clarify the nomenclature we have named the corresponding allele rad51d-3 with rad51d-1 and rad51d-2 being the alleles previously characterised by Durrant et al. (2007) and Inagaki et al. (2009) respectively. Plants homozygous for rad51d-3 T-DNA insertions were identified by PCR in the T3 seeds provided by the Nottingham Arabidopsis Stock Centre. As for other xrcc2 and rad51b mutants, no obvious growth or fertility defects were observed in plants homozygous for rad51d-3. Molecular characterisation of the insertion was done by amplifying the T-DNA junctions and sequencing of the corresponding PCR products. The results are summarised in panel (A). The rad51d-3 allele T-DNA is inserted in exon 8 of the RAD51D gene and the insertion is associated with a deletion of 11bp of the exon. As frequently observed, the inserted T-DNA is flanked by two left borders in opposite orientations. Exons are shown as boxes and PCR primers as arrowheads. (B) Plants homozygous for the T-DNA insertion were selected by PCR, and semi-quantitative RT-PCR analysis performed to assess the presence of AtRAD51D transcripts in total RNA isolated from wild-type and mutant leaves. AtRAD51D transcripts were detected in the wild-type but no AtRAD51D mRNA could be detected using primers flanking the T-DNA insertion in RNA from rad51d-3 plants. In contrast, truncated AtRAD51D mRNAs could be detected upstream of the T-DNA insertion. The T-DNA insertion in the atrad51d-3 mutants thus prevents the production of the full-length mRNAs (and hence proteins) of AtRAD51D. (C). Mutation in Arabidopsis RAD51D confers hypersensitivity to the DNA cross-linking agent, Mitomycin C (MMC). Percentages of wildtype and rad51d sensitive plants at different MMC doses (0, 20, and 40 µM MMC). 50 plantlets were analysed in each case. In the absence of MMC, wild-type plants develop at least four true leaves (excluding the cotyledons), thus plants with three leaves or less were considered as sensitive. Figure S2. Crossing scheme for the introgression of Landsberg markers into the xrcc2 mutant background. Landsberg erecta plants were crossed with xrcc2 mutants (Col0 background). F2 plants were tested for heterozygosity of the markers and XRCC2 genotype. Marker segregation analyses were done on progeny of these F2 plants. Table S1. List of PCR markers between Col and Ler. For each marker, the corresponding Bacterial Artificial Chromosome, primers, size of the Columbia and Landsberg products, and the physical distance of the intervals are listed.