jam12831-sup-0001-TableS1-S7
advertisement
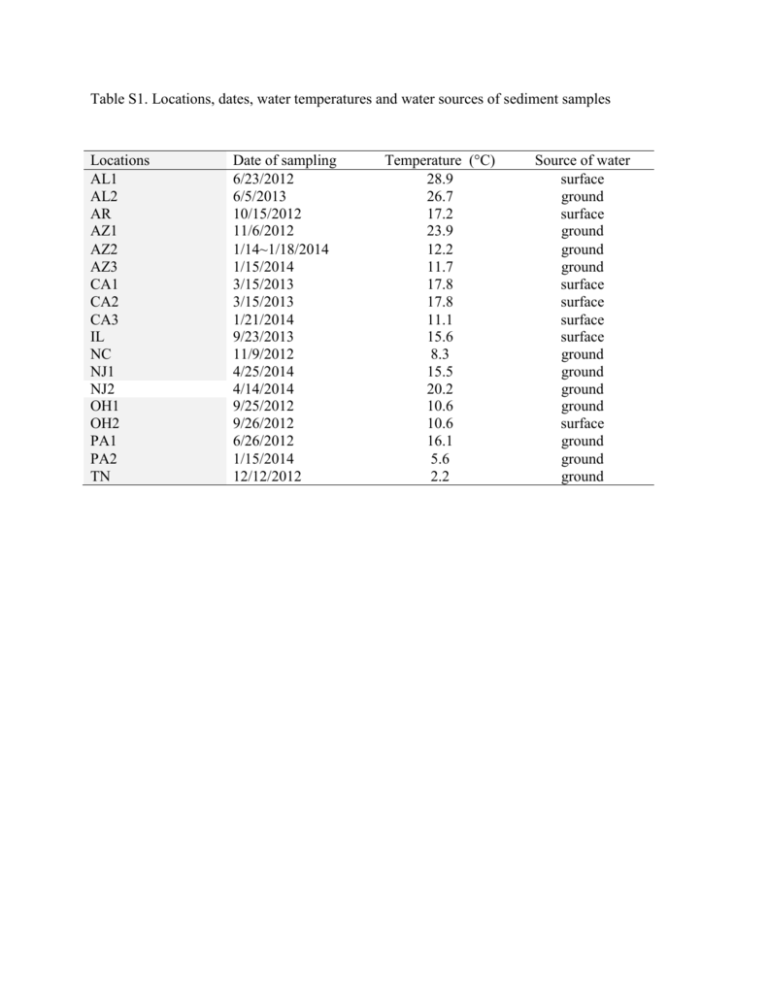
Table S1. Locations, dates, water temperatures and water sources of sediment samples Locations AL1 AL2 AR AZ1 AZ2 AZ3 CA1 CA2 CA3 IL NC NJ1 NJ2 OH1 OH2 PA1 PA2 TN Date of sampling 6/23/2012 6/5/2013 10/15/2012 11/6/2012 1/14~1/18/2014 1/15/2014 3/15/2013 3/15/2013 1/21/2014 9/23/2013 11/9/2012 4/25/2014 4/14/2014 9/25/2012 9/26/2012 6/26/2012 1/15/2014 12/12/2012 Temperature (°C) 28.9 26.7 17.2 23.9 12.2 11.7 17.8 17.8 11.1 15.6 8.3 15.5 20.2 10.6 10.6 16.1 5.6 2.2 Source of water surface ground surface ground ground ground surface surface surface surface ground ground ground ground surface ground ground ground Table S2. Sediment parameters across seven sampling sites Particle Size Total Tank Exchange pH % Location % Clay Capacity % Silt Sand (ME/100g) TOC (%) Organic Matter (%) Source water TN 0.4 1.06 98.54 8.2 3 0.42 0.43 ground NC 7.63 23.39 68.98 7.6 110.8 3.11 5.45 ground OH1 1.36 6.7 91.94 7.1 26.7 0.25 0.89 ground AL1 2.73 2.33 94.94 7.8 12.2 0.42 0.88 surface AR 3.91 14.44 81.65 6.7 12.17 2.78 11.45 surface OH2 1.68 21.67 76.65 6.6 57.34 2.09 5.9 surface AZ1 7.35 34.34 58.31 6.7 154.14 9.42 4.08 ground Table S3. Screening analyses and methods performed on sediment samples Test parameter Method Total organic carbon Automated instrumental analysis of carbon and nitrogen in plant and soil samples (1) (Comparable to EPA Method 9060A) (2). Estimation of soil organic matter by weight, loss on ignition (3) (Comparable to EPA Method 160.4 (4). ASTM D422 (5) (sieve/hydrometer) EPA Method 9045C rev 3 (6) Total organic matter Particle size analysis (sand, silt, and clay) pH Total exchange capacity U.S. EPA, EPA Method 9080, Cation Exchange Capacity in Soils, Rev. 0, 1986 (7). References 1. McGeehan, S.L., and D.V. Naylor. 1988. Automated instrumental analysis of carbon and nitrogen in plant and soil samples. Commun. Soil Sci. Plant Anal. 19:493-505. 2. U.S. EPA, EPA Method 9060A, Total Organic Carbon, Rev. 1, November 2004. 3. Schulte, E.E., and B.G. Hopkins. 1996. Estimation of soil organic matter by weight LossOn-Ignition. P. 21-32; in: Soil Organic Matter: Analysis and interpretation. (ed.) F.R. Magdoff, M.A. Tabatabai, and E.A. Hanlon, Jr. Special publication No. 46. Soil Sci. Soc. Am. Madison, WI. 4. U.S. EPA, EPA Method 160.4, Volatile Residue, Rev. 1, 1971 in Methods for Chemical Analysis of Water and Wastes, EPA/600/4-79/020, March 1983. 5. ASTM Standard D422, 1998, “Standard Test Method for Particle-Size Analysis of Soils,” ASTM International, West Conshohocken, PA, www.astm.org. 6. U.S. EPA, EPA Method 9045C, pH in Liquid and Soils, Rev. 1, 2000. 7. U.S. EPA, EPA Method 9080, Cation Exchange Capacity in Soils, Rev. 0, 1986. 8. Table S4. Oligonucleotide taqMan qPCR primer sequences used to screen pathogens and humanfecal indicator Bacteroidetes and PCR primer sequences for clone sequencing Target Human-specific HF183 Bacteroidales (qPCR) Campylobacter spp. (qPCR) Oligo name Sequences (5'-3') Tm (°C) HF183F ATCATGAGTTCACATGTCCG 60 BthetP1 FAMCTGAGAGGAAGGTCCCCCACATTG GA-TAMRA CGTAGGAGTTTGGACCGTGT BthetR1 CampF2 CampR2 CampP2 Campylobacter spp. (PCR) Legionella spp. (qPCR) CampF CampR Leg F1c Leg R1c Legprobe Legionella spp. (PCR) LegF LegR1 LegR3 mipF1a L. pneumophila (qPCR) mipR1 mipProbe L. pneumophila (CYBR-Green qPCR) L. pneumophila (CYBR-Green qPCR) L. pneumophila (CYBR-Green qPCR) L. pneumophila (CYBR-Green qPCR) L. pneumophila (qPCR) rpsLf rpsLr rtxAf rtxAr sidFf sidFr cegC1-F cegC1-R P65f P66r CACGTGCTACAATGGCATAT GGCTTCATGCTCTCGAGTT FAMCAGAGAACAATCCGAACTGGGACA -TAMRA GGA TGA CAC TTT TCG GAG C CAT TGT AGC ACG TGT GTC TAGTGGAATTTCCGGTGTA CCAACAGCTAGTTGACATC 6FamCGGCTACCTGGCCTAATACTGATamra GATTAGCCTGCGTCCGATTAG GAAATTCCACTACCCTCTCCCA AGTGTCAGTATTAGGCCAGGTAGC AGGATAAGTTGTCTTATAGCA TTAAGAACGTCTTTCA TTTG 6FAMTAATCCGGAAGCAATGGCTAATamra GAAAGCCTCGTGTGGACGTA CAACCTTACGCATAGC TGAGTTA ATTGCGCCTGGCAAACTTTAGGTG GGCGCAAATCGTTTCCACCTTGTA ATTGTTCGCGAGGGTATGAAAGC G TCTTTCCAAGACAGACTCTCGCGT TGCCTAAACGGTATGACCGCATCA GGCATATGCACCAAAC CACCGAAT CAAAGGGCGTTACAGT CAAACC CAAACACCCCAACCGT AATCA Detection limit (CE per reaction) 2 Reference Bernhard and Field (2000) Shanks et al. (2008) Converse et al. (2009) 58 1 Lund et al. 2004 60 2 Linton, 1996 50 2 This study 64 2 This study 60 2 This study 64 2 Lu, et al. 2013 64 2 Lu, et al. 2013 64 2 Lu, et al. 2013 64 2 Faucher et al. 2011 60 2 Lp-lg1,75bp, 60C, Mérault 2011 Salmonella spp. (qPCR) Mycobacterium spp. (ID) Mycobacterium spp. (qPCR) sg1-pb invA_176 F invATx_208 invA_291 R T-39 T-13 23Smyco F 23Smyco R 23Smyco Probe FAMTCTTGGGATTGGGTTG GGTTATTTTAACTCCTBHQ CAACGTTTCCTGCGGT ACTGT FAMCTCTTTCGTCTGGCATTATCGATCA GTACCA-TAMRA CCCGAACGTGGCGATAATT TGCACACAGGCCACAAGGGA GGG GTGTGGTGTTTGAG 62 1 Bruijnesteijn van Coppenraet, 2004 60 2 Riviere et al. 2006 56 2 Kuiper et al. 2006 60 2 Schroeder et al. 2001 60 2 Hadfield et al 2011 60 2 Hill et al. 2007 60 2 Guy, 2003 63 1 Qvarnstrom et al. 2006 Vermamoeba vermiformis (qPCR) Acanthamoeba spp. (PCR) Hv1227F Hv1728R JDP1 GGCCCAGATCGTTTACCGTGA A JDP2 CRU18Sf TCTCACAAGCTGCTAGGGAGTCA GAG GTA GTG ACA AGA AAT AAC AAT ACA GG CTG CTT TAA GCA CTC TAA TTT TCT CAA AG 6FAM-TAC GAG CTT TTT AAC TGC AAC AA-BHQ ATG ACG GGT AAC GGG GAA T CCA ATT ACA AAA CCA AAA AGT CC CGC GCC TGC TGC CTT CCT TAG ATG CATCCGCGAGGAGGTCAA JVA18S f r p Giardia spp. qPCR Naegleria fowleri β-Giardin P241 F R P NaegIF19 2 NaegIR3 44 NfowlP Talaat et al. (1997). CTCCCACGTCCTTCATC TaqAcF1 TaqAcR1 TaqAcP1 P GonzalezEscalona, N., et al. 2009 50 Acanthamoeba spp. (qPCR) r 4 GCGAACGGGTGAGTAACACG 6-carboxyfluoresceinTGGATAGTGGTTGCGAGCATCBHQ1 CGACCAGCGATTAGGAGACG CCGACGCCAAGGACGAC FAMTGAATACAAAACACCACCATCGGC GC-TAMRA TTA CGA GGT CAG GAC ACT GT GAC CAT CCG GAG TTC TCG Cryptosporidium spp. (qPCR) 60 GCAGCCATGGTGTCGATCT FAM/AAGTCCGCCGACAACATGTA CCTAACGA/BHQ-1 GTG CTG AAA CCT AGC TAT TGT AAC TCA GT CAC TAG AAA AAG CAA ACC TGA AAG G HEX-AT AGC AAT ATA TTC AGG GGA GCT GGG C-BHQ1 References Bernhard A.E. and Field, K.G. (2000) A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574. http: //dx.doi.org/10.1128/AEM.66.10.4571-4574.2000. Converse, R.R., Blackwood, A.D., Kirs, M., Griffith, J.F. and Noble, R.T. (2009) Rapid QPCRbased assay for fecal Bacteroides spp. as a tool for assessing fecal contamination in recreational waters. Water Res 43, 4828–4837. Lu, J., Struewing, I., Buse, H.Y., Kou, J., Shuman, H.A., Faucher, S.P. and Ashbolt, N.J. (2013) Legionella pneumophila transcriptional response following exposure to CuO nanoparticles. Appl Environ Microbiol 79: 2713-2720. Gonzalez-Escalona N., et al. (2009) Detection of live Salmonella sp. cells in produce by a TaqMan-based quantitative reverse transcriptase real-time PCR targeting invA mRNA. Appl Environ Microbiol 75:3714–3720. Talaat, A.M., Reimschuessel, R. and Trucksis, M. (1997) Identification of mycobacteria infecting fish to the species level using polymerase chain reaction and restriction enzyme analysis. Vet Microbiol 58:229-37. Bruijnesteijn van Coppenraet, E.S., Lindeboom, J.A., Prins, J.M., Peeters, M.F.,E. Claas, C.J. and Kuijper, E.J. (2004) Real-time PCR assay using fine-needle aspirates and tissue biopsy specimens for rapid diagnosis of mycobacterial lymphadenitis in children. J Clin Microbiol 42:2644 Riviere, D., Szczebara, F.M., Berjeaud, J.M., Frere, J. and Hechard, Y. (2006) Development of a real-time PCR assay for quantification of Acanthamoeba trophozoites and cysts. J Microbiol Methods 64, 78–83. Schroeder, J.M., Booton, G.C., Hay, J., Niszl, I.A., Seal, D.V., Markus, M.B.,Fuerst, P.A. and Byers, T.J. (2001) Use of subgenic 18S ribosomal DNA PCR and sequencing for genes and genotype identification of acanthamoebae from human with keratitis and sewage sludge. J Clin Microbiol 39:1903–1911. Hadfield, S.J., Robinson, G, Elwin, K., Chalmers, R.M. (2011) Detection and differentiation of Cryptosporidium spp. in human clinical samples by use of realtime PCR. J Clin Microbiol 49: 918–924. Hill, V.R., Kahler, A.M., Jothikumar, N., Johnson, T.B., Hahn, D., et al. (2007) Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl Environ Microbiol 73: 4218–4225. Qvarnstrom, Y., Visvesvara, G.S., Sriram, R. and da Silva, A. J. (2006). Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J Clin Microbiol 44, 3589-3595. Table S5. Legionella strains used to test PCR coverage Name ATCC # Designation Source Assay (F2/R1) Legionella cincinnatiensis 43753 72-OH-0 human lung tissue, Cincinnati, OH + Legionella dumoffii 33279 NY23 Cooling tower, New York + Legionella feeleii 35849 91-WI-H, human lung tissue, Wisconsin + Legionella gormanii 33297 LS-13 [ALLO3], soil from creek bank, Atlanta, GA + Legionella hackeliae 35250 Lansing 2 human bronchial biopsy, Ann Arbor, MI + Legionella jamestowniensis 35298 JA-26-G1-E2 wet soil, Jamestown, NY + Legionella longbeacheae 33462 Long Beach 4 human lung + Legionella maceachernii 35300 PX-1-G2-E2 water in home evaporator cooler, Phoenix, AZ + L. micdadei [Tatlockia micdadei] 33218 TATLOCK blood-human (human blood via yolk sac) + Legionella oakridgensis 33761 Oak Ridge 10 industrial cooling tower + water, PA Legionella quinlivanii 43830 1442-AUS-E water in bus air conditioner, Austrailia + Legionella rubrilucens 35304 WA-270A-C2 tap water, Los Angeles, CA + Legionella sainthelensis 35248 MSH-4 spring water, Mt. St. Helens, WA + Legionella wadsworthii 33877 L. pneumophila strain Lp02 Wadsworth 81- sputum, South Carolina 716A Dept of Health and Environmental Control + n/a + L. pneumophila subsp. pneumophila strain Philadelphia-1 33152 human lung + L. pneumophila subsp. pneumophila strain Bloomington-2 33155 creek water + L. pneumophila subsp. fraseri strain Dallas 1E 33216 cooling tower + L. pneumophila; subsp. pneumophila strain Chicago 2 33215 human lung biopsy + Table S6 Bacterial strains used to check the specificity of the PCR and qPCR assays for Legionella Strains Source Strains Source Acinetobacter baumannii ATCC 19606 Micrococcus luteus ATCC 10240 Aeromonas hydrophila ATCC 7966T Proteus mirabilis ATCC 12453 Aeromonas hydrophila ATCC 7966 Proteus vulgaris ATCC 29905 Aeromonas cavae ATCC 15468 Pseudomonas aeruginosa ATCC 10145 Bacillus cereus ATCC 10876 Salmonella enteriditis ATCC 13076 Burkholderia cepacia ATCC 25416 Serratia marcescens ATCC 14756 Burkholderia cepacia ATCC 25416 Serratia marcescens ATCC 13880 Campylobacter jejuni ATCC 29428 Shewanella putrefaciens ATCC 49138 Citrobacter freundii ATCC 4391 Shigella sonnei ATCC 25931 Citrobacter freundii ATCC 8090 Shigella sonnei ATCC 9290 Enterobacter aerogenes ATCC 13048 Staphylococcus aureus ATCC 25923 Enterobacter aerogenes ATCC 13048 Staphylococcus aureus ATCC 25923 Enterobacter cloacae ATCC 13047 Streptococcus pyogenes ATCC 19615 Enterococcus faecalis ATCC 29302 Yersinia enterocolitica ATCC 23715 Enterococcus faecalis ATCC 19433 Escherichia coli EPA isolate 8 Enterococcus faecium ATCC 19434 E. coli EPA isolate AD#1 Lactobacillus acidophilus ATCC 314 E. coli EPA isolate 58 Klebsiella oxytoca ATCC 13182 E. coli EPA isolate 59 Klebsiella pneumoniae ATCC 13882 E. coli EPA isolate 62 Klebsiella pneumoniae ATCC 31488 Legionella pneumophila strain Lp02 33152 Listeria monocytogenes Scott O2 L. pneumophila subsp. pneumophila strain Philadelphia-1 Table S7. Total clone numbers, unique OTUs and dominant sequences in those sediments showing positive for Legionella spp. Site AL1 Total clone 67 OTU# 6 AZ1 23 7 AZ2 24 4 IL NC NJ1 58 25 62 16 12 36 NJ2 36 9 OH2 17 13 PA2 TN 9 15 6 6 Dominant L impletisoli str OA1 1 (AB233209) unknown L impletisoli str OA1 1 (AB233209) unknown L impletisoli str OA1 1 (AB233209) L adelaidensis (Z49716) unknown unknown unknown L pneumophila subsp pneumophila str Philadelphi L anisa strain L rubrilucens L rubrilucens L brunensis % 22 72 30 26 29 21 17 93 64 2 31 42 64 11 L impletisoli L cincinnatiensis L brunensis S. lyticum L pneumophila subsp pneumophila str Philadelphi 18 12 18 89 100