SCLS-2015-0189.R2 Method_Online
advertisement

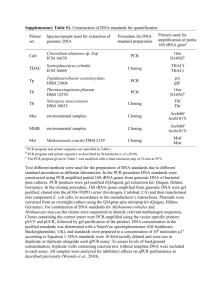
Method Online Sample collection and DNA extraction Thirty commercial products of yak jerky were purchased from commercial markets in four cities of China, including Linzhi, Tibet (n=2), Chengdu, Sichuan (n=8), Kunming, Yunnan (n=11) and Xining, Qinghai (n=9) (Fig 1). In addition, three commercial products of beef jerky were purchased in Kunming and work as controls. Details of all samples were summarized in Table S1. All samples were stored at -40 ℃ before further processing. 10 mg of each jerky sample was rinsed with distilled water overnight and used for genomic DNA extraction using the standard phenol/chloroform method. PCR amplification and sequencing The standard COI-5P fragment was amplified using the universal primers described by Che et al. [1] (Forward: COI-C01: TYTCWACWAAYCAYAAAGAYATTGG and COI-C02: AYTCAACAAATCATAAAgATATTGG; Reverse: COI-C03: ACYTCYGGRTGACCAAARAAYCA and COI-C04: ACYTCRGGRTGACCAAAAAATCA). A fragment of 16S was also amplified using 16SAR (5'-CGCCTGTTTAYCAAAAACAT-3') and 16SBR (5'CCGGTYTGAACTCAGATCAYGT-3') [2]. The PCR amplification reaction was performed in a total volume of 25 µL, including 1µl (50 ng) of genomic DNA, 17.5µl of ddH2O, 2.5µl of 10 X PCR buffer, 1.0µl (10 pmol) of each primer, 1.0µl (0.1mg⁄mL) of BSA, 1.0µl (2.5mM) of dNTP and 0.12µl (5U/µl) of Taq polymerase. The PCR conditions were the following: an initial denaturation step with 5 min at 95℃, then 35 cycles of denaturation 1 min at 94 ℃, annealing for 1 min at either 45 ℃ for COI or 55 ℃ for 16S, extension for 1 min at 72℃. Final extension at 72 ℃ was conducted for 10 min. Species identification and data analysis Sequencing was performed in both directions using BigDye Terminator Cycle Sequencing Kit (version 3.1, Applied Biosystems) and an ABI 3730xl genetic analyzer (Applied Biosystems). All amplified sequences were manually edited using Geneious R6 (Biomatters). The nucleotide sequences were translated into amino acids to discover premature stop codon, i.e. to exclude the sequences of nuclear DNA pseudogenes. We used both IDS engine of BOLD system (www.boldsystems.org) [3] and NCBI Blast (blast.ncbi.nlm.nih.gov/Blast.cgi) to identify species (Table S1). A tree-based identification method was also applied for more reliable species identification. All COI and 16S sequences including reference sequences downloaded from BOLD or GenBank (Standard Barcodes) were aligned using ClustalX 1.81 [4] with default parameters and then manually optimized in MEGA 4.0 [5]. The neighbor-joining (NJ) trees based on COI and 16S were built in MEGA 4.0 with the default parameters (Fig. 1). Robustness of the trees was tested using 1,000 bootstrap replicates. According to the identification results, a True-False table was calculated (Fig. 1). References: [1] Che J, Chen H-M, Yang J-X, Jin J-Q, Jiang KE, Yuan Z-Y, Murphy RW, Zhang Y-P. Universal COI primers for DNA barcoding amphibians. Molecular Ecology Resources, 2012, 12: 247-258 [2] Palumbi SR. Nucleic acids Ⅱ: the polymerase chain reaction. In: MABLE DMHCMMNK, eds. Molecular systematics, 2nd ed. Sunderland, Massachusetts, USA: Sinauer Associstes, Inc, 1996. 205-247 [3] Ratnasingham S, Hebert PDN. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes, 2007, 7: 355-364 [4] Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Research, 1997, 25: 4876-4882 [5] Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution, 2007, 24: 1596-1599