Supplemental Digital Content 1
advertisement

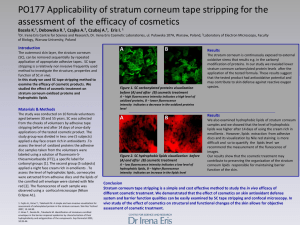
Supplemental Digital Content 1 Methods LAURDAN Generalized polarization function For the LAURDAN GP measurements the fluorescence signal were collected on the microscope in two different channels simultaneously. The experimental setup is described in detail in[1]. The fluorescence signal was split between two PMTs (Hamamatsu H7422P40) by a dichromatic mirror splitting at 475 nm. The PMTs had bandpass filters mounted 438 12nm and 494 10 nm respectively. LAURDAN generalized polarization (GP) measurements [2] were used to detect and evaluate the organization of the extracellular lipids within the cholesteatoma and the normal skin stratum corneum, as described by [3]. LAURDAN is an amphiphilic probe that preferentially partitions into lipid membranes. The fluorescence emission properties are sensitive to the water content and dynamics that occurs in the probe’s environment. Briefly, the energy of the LAURDAN excited singlet state progressively decreases when the extent of water dipolar relaxation process is augmented, red shifting the probe’s emission spectrum, i.e. decreasing the GP function to lower values (see equation 1). The extent of water dipolar relaxation is normally related to the lipid packing of the studied membrane being low when the membrane packing is tight (e.g. membranes in the gel phase) [1]. Likewise, LAURDAN GPs were calculated using the formula: GP I B G I R I B G I R (1) where IB and IR correspond to the intensities at the blue and red edges of the emission spectrum (440 and 490 nm) using a given excitation wavelength [1, 4]. In lipid bilayers high LAURDAN GP values (0.5 – 0.6) correspond to laterally ordered phases (e.g solidordered or gel) whereas low LAURDAN GP values (below 0.1) correspond to liquiddisordered -like phases [1, 2]. The correction factor G was calculated by acquiring GP images of a known LAURDAN reference solution in the microscope at the same instrumental conditions used in the tissue sections (for further details see [1]). LAURDAN (2µM) in DMSO was used, with the predetermined GP=0.011. The reference’s GP was measured as described [1]. Lipid extraction and lipid analyses In these experiments, the lipids were extracted from10 individual cholesteatoma samples. The control samples obtained from normal human skin stratum corneum (see materials section) were collected from at least 6 individuals. Samples of human skin stratum corneum from breast reduction surgery were isolated after overnight floating on 1% trypsin solution (cell culture grade). Cells from the viable epidermis were removed from stratum corneum sheets by repeated rigorous shaking. The cholesteatomas were rinsed thoroughly in 0.9% NaCl (by weight) immediately after surgery to avoid contamination with blood. Each sample preparation was checked under the microscope to ensure lack of viable epidermal cells. All samples, i.e. cholesteatoma and human skin stratum corneum were rinsed in 0.88% KCl (by weight) prior to homogenization and homogenized for 3 x 1 minutes using an UltraThurrax® at max speed. Samples were dried and weighed. Lipids were extracted by a modified Bligh and Dyer [5] method: Dry tissue was suspended in a total volume of 1.9mL chloroform:methanol:0.88% KClaq 2:1:0.8 (by volume). Following rigorous mixing and sonication, the ratio of solvents was changed to chloroform:methanol:0.88% KClaq 1:1:0.9 (by volume), and samples were again rigorously mixed. Following centrifugation the solution divides into an upper aqueous phase and a lower organic phase. The lower (organic) phase was transferred to a new tube and the upper (aqueous) phase was re-extracted twice with fresh solvent from the lower phase of the chloroform:methanol:0.88% KClaq 1:1:0.9 (by volume) mixture. The combined lower phases were washed once with fresh upper phase from the chloroform:methanol:0.88% KClaq 1:1:0.9 (by volume) mixture and dried under a stream of nitrogen. Lipids were resuspended in the lower phase of the chloroform:methanol:0.88% KClaq 1:1:0.9 (by volume) mixture and filtered through a solvent resistant 0.45µm filter prior to analysis. High performance thin layer chromatography (HPTLC) analyses: Lipid extracts were diluted according to dry tissue weight and extracts from individual samples corresponding to 150µg starting material (tissue dry weight) was loaded onto the silica plates under a stream of nitrogen. Plates were developed with chloroform-acetonemethanol 76:4:20 (first up to 10 mm, followed by up to 25 mm), then chloroform-acetonemethanol 80:10:10 (up to 75 mm) as the second, and chloroform-diethyl ether-ethyl acetate-methanol 72:6:20:2 (up to 90 mm) as the third developing system. Cholesterol containing lipids were visualized by staining plates with 10%FeCl3 in 5% CH3COOH and 5% H2SO4 (pink/purple for cholesterol) followed by baking at 100°C for 2-7 minutes. Remaining lipids on the plates were stained with 10% CuSO4 in 8% H2PO3 followed by baking at 180°C for 10-15 minutes. Standards included for identification of co-migrating bands were: Cholesterol and cholesterol sulphate (SigmaAldrich), Nu-Chek 18-4A standard (Nu-Chek Prep, Inc.; which contains equal amounts (by weight) of cholesterol, oleic acid, methyl olein, triolein, cholesteryl ester), and a free fatty acid mix (FFA) contain equal amounts (by weight) of C19-C26 fatty acids acquired separately from Larodan Sweden. 1. 2. 3. 4. 5. Brewer, J., et al., Multiphoton excitation fluorescence microscopy in planar membrane systems. Biochim Biophys Acta, 2010. 1798(7): p. 1301-8. Bagatolli, L.A., To see or not to see: lateral organization of biological membranes and fluorescence microscopy. Biochim Biophys Acta, 2006. 1758(10): p. 1541-56. Carrer, D.C., C. Vermehren, and L.A. Bagatolli, Pig skin structure and transdermal delivery of liposomes: a two photon microscopy study. J Control Release, 2008. 132(1): p. 12-20. Parasassi, T., et al., Laurdan and Prodan as Polarity-Sensitive Fluorescent Membrane Probes. Journal of Fluorescence, 1998. 8(4): p. p362-373. Bligh, E.G. and W.J. Dyer, A rapid method of total lipid extraction and purification. Can J Biochem Physiol, 1959. 37(8): p. 911-7.