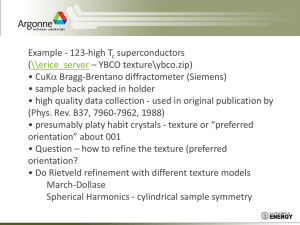
Fitequipoint3D manual V 1.0
advertisement

Fitequipoint3D/FEP3D manual
a software to visualize reciprocal space
By Simon Gaudet
Foreword
The goal of this manual it to help user who are not familiar with the fitequipoint program to use
it, to help introduced user perform more advanced task and to allow advanced user to modify
the code to their own needs. I hope this small manual help you solve interesting material
problem. Video tutorial complementary to this manual are also available.
Table of Contents
Foreword ......................................................................................................................................... 2
Table of Content ................................................................................ Error! Bookmark not defined.
1
Introduction ............................................................................................................................. 4
1.1
What is reciprocal space? .................................................. Error! Bookmark not defined.
1.2
What is a pole figure? ........................................................ Error! Bookmark not defined.
1.3
What information can we get out of this? ........................ Error! Bookmark not defined.
2
Installing the program ............................................................................................................. 6
3
The interfaces .......................................................................................................................... 7
4
Basic operation ........................................................................................................................ 9
5
6
4.1
Loading data .................................................................................................................... 9
4.2
Loading crystal ................................................................................................................. 9
4.3
Modifying crystal ........................................................................................................... 10
4.4
Plotting pole figure ........................................................................................................ 11
Fitting an orientation ............................................................................................................. 12
5.1
Epitaxial texture............................................................................................................. 12
5.2
Fitting fiber texture on pole figures .............................................................................. 13
5.3
Fitting axiotaxial texture on pole figures....................................................................... 13
Scanning through reciprocal space ....................................................................................... 17
6.1
Performing reciprocal space cuts .................................................................................. 18
1 Introduction
X-ray-diffraction enables us to measure full volumes of reciprocal space of a sample,
whether a single-crystal, a large number of grains of the same material, or even a mixture of
different phases. The goal of the software explained in this manual is to navigate through
reciprocal space to characterize the nature and orientations of grains of a material. Each grain
can be associated with a reciprocal space representation as shown, for example, in Fig. 1(a) for a
Ni crystal. The reciprocal-space image of a sample is the overlap of the reciprocal lattices of the
individual grains. Thus, the reciprocal image depends, in the case of polycrystalline materials, on
texture – on the orientation of all grains in the sample.
Figure 1: Schematic representation of the reciprocal space of a face-centered crystal (such as Ni) for (a) an epitaxial
texture, (b) a fiber texture, and (c) a randomly oriented film.
Four types of texture components are usually considered in the thin film literature:
(i) random texture, where the grains have no preferential orientation; (ii) fiber texture, for which
one of the crystallographic axis of the film is parallel to the substrate normal while there is a
rotational degree of freedom around the fiber axis; (iii) epitaxial alignment, when an in-plane
film alignment fixes all three axis with respect to the substrate, and (iv) axiotaxy, a type of
texture characterized by the alignment of planes in the film and substrate that share the same
interplanar spacing.i This preferred alignment of planes at the interface manifests itself as a fiber
texture lying off-normal to the sample surface, with the fiber axis perpendicular to the planes
that are aligned at the interface.
For an epitaxial texture, all grains have the same orientation; consequently, the reciprocal image
is also a single lattice as shown in Fig. 1(a). In the case of a fiber texture, the rotational degree of
freedom around the fiber axis leads to a reciprocal image composed of rings centered on this
axis, as represented in the schematics of Fig. 1(b). For a random texture, all grain orientations
are equally probable; the reciprocal image is thus a series of concentric shells centered on the
origin of the reciprocal space (Fig. 1(c)). Axiotaxy will appear in reciprocal space as an off-normal
fiber texture. As a final point, it should be understood that these 4 texture types can appear in a
film of single or mixed phases. In a sample with mixed phases, each phase could be represented
by more than one texture components and the final reciprocal image is the overlap of the
reciprocal images of each of the phases.
In the rest of this manual we will show the user how to load the data, look at them in order to
identify the different phases and texture.
2 Installing the program
1) Unzip the folder containing the program on your hard drive
2) Make sure that the office web components 11.0 are install on your hard drive. If this
component is not installed you will not be able to change the crystal structure of your
material. At this point this feature is only supported for windows and not for Mac. To
install the web office component download the owc11.exe file from the knowledge
base
website
of
Microsoft
and
run
the
executable:
http://www.microsoft.com/downloads/en/details.aspx?FamilyId=7287252C-402E-4F7297A5-E0FD290D4B76&displaylang=en
3) Start Matlab
4) Add the path of the program to the Matlab path by selecting in the upper menu : file
Set path… and by selecting Add with subfolder… and choosing the root path of the
decompresses file
5) run the program by typing the latest version name in the command line of Matlab
(fitequipoint3d12 or FEP3D13)
3 The interfaces
4 Basic operation
4.1 Loading data
By clicking on “Load XRD” a user interface pops-up and ask you to load a file. This file is a .mat
file that contains a structure name temp.
Data structure:
This structure must contain the following sub-variable:
temp.lambda : the wavelength;
temp.tth_min: the minimum 2
temp.tth_nb: the numbers of 2 considered
temp.theCELL: a big structure containing all the information
“temp.theCELL” is a structure of the size temp.tth_nb x 7. Each element can have a different
size. In matlab such element are referred using {} for example temp.theCELL{1,1}. For each
row a different 2 is used. The seven column are:
1: 1x3 matrix [tth Iavg nbpix]
tth= 2
Iavg=the average intensity at that 2 LEGACY
nbpix=the number of pixel that contributed to the intensity LEGACY
2: nb_psi x nb_phi matrix. All lines have different psi but on a line they are all the same. This is
used to plot pole figure as the variable psi
3:nb_psi x1 ; the summed intensity for each at the specified d-spacing LEGACY
4:nb_psi x1: the number of pixel that contributed to the previous intensity LEGACY
5: nb_psi x nb_phi matrix. All lines have the same value of phis but on a line they are different
This is used to plot pole figure as the variable
6: nb_psi x nb_phi matrix. The normalized intensity for each specified and
7: nb_psi x nb_phi matrix. The number of pixel that contributed to each intensity LEGACY
The variable identified as LEGACY are only used by another software PoFiAD (also called
respad)
4.2
Loading crystal
By clicking on “Load crystal” a pop-up open and allows loading a crystal. There are two
possibility either .cry or .cvs. The .cry makes reference to the format used by HKL (for EBSD). The
.cvs is a format created for this program.
4.3 Modifying crystal
To modify a crystal click on “detailed film properties”. The information about the crystal is then
displayed with the a,b and c lattice constant as well as the , gamma angles. Each atom
position in the lattice is specified. The atomic number also needs to be specified. This number is
latter used to determine the atomic scattering factor. The radius and color are used only in the
visualizes3D2 application that allows you to see the crystal structure. The occupancy (the
percentage of occupation of a site) is used to calculated peak relative intensity. If this applet
does not load, it might be that you have not installed the office web component 2003 on your
pc.
http://www.microsoft.com/downloads/en/details.aspx?familyid=7287252C-402E-4F7297A5-E0FD290D4B76&displaylang=en (On Mac it does not work. A new file format directly
editable in finder should be programmed at some point for Mac user.).
To apply the modification click Save value.
To save for later use Save crystal.
To create a new crystal just modify an old one and save it with a new name.
4.4 Plotting pole figure
Prior to plotting a pole figure a crystal must
have been selected (1), and data must have
been loaded (2).
The user must also select a pole figure to
plot (3) using the checkboxes. Up to three
pole figures can be plotted on selected
Matlab figures (4).
There are two ways to plot pole figures:
1) Using planes of the define crystal (5)
2) Using the d-spacing (6)
To use the d-spacing the use d check box,
(7), need to be checked, otherwise the
planes of the crystal will be used.
(1)
(2)
(9)
(12)
(10)
(7)
The pole figure plotted is the summation of
the different pole figure near the selected d
or plane. The range is selected using the
delta d edit boxes (8).
(11)
(5)
To plot a pole figure you need to click on
Plot pole figures (9).
(3)
(4)
(8)
(6)
The pole figures can be plotted with vairous
symetry using the combo box (10). It will
work only for data where empty region are
not included in the structure but not if zeros
are included (as in RESPAD software result).
The log of the results will be plotted if the
Log graph checkbox is checked. (11)
The color minimum and maximum also need
be change to fit the data scale (12). Data need to be replotted to modify the scale. The user can
also used Matlab built in function such as colormap and caxis.
5 Fitting an orientation
5.1 Epitaxial texture
To fit an orientation of a specific
crystal you need to first select the
crystal you want the fit as explained
before.
Based on a plausible hypothesis for
the nature of the phase, we select a
feature corresponding to a specific
(hill) set of values.
All rotations applied to the
simulated crystal are performed in
respect to a plane that is selected (1)
by the user by clicking on Simul.
Epitaxie (2).
By default, this plane is considered
to be parallel to the surface. We
must first apply a rotation to bring
this plane in coincidence with the
observed feature. A (3)and (4)
rotation changes the orientation of
the entire crystal; therefore, all
simulated diffraction features move
accordingly, but on the pole figure of
the specified plane it will move the
specified plane to the and of the
pole figure.
(2)
(6)
(10)
(1)
(8)
(4)
(3)
(7)
(9)
(5)
The final degree of freedom in the
crystal position consists in a rotation
around the selected plane surface
normal (2) (5). All other fitted peaks
will then rotate around the selected
plane. To see all the possible
positions of the peaks using this rotation the user can click Simul Fiber (6). If the peak
identification is correct, a judiciously selected value of 2 must therefore lead to a perfect
agreement for all peak positions and intensities. If more than one phase or orientation is
present, it is possible that diffraction peaks overlap, thereby modifying the observed intensities.
In order to facilitate the comparison of the experimental and calculated intensities, the opacity
of the square corresponding to each diffraction plane is proportional to its relative intensity; the
most intense peaks being opaque and the forbidden peaks completely transparent. The opacity
of the square edges corresponds to the square root of the relative intensity and they are
therefore always slightly more visible than the center.
It is critically important to insist on the fact that diffraction peaks must fit not one but all pole
figures self-consistently. The current software can overlay the fitted orientations in a video of
pole figures which allows for a rapid evaluation of the quality of the fit over the full reciprocal
space. It is also possible to overlay the fit of any crystal orientation on selected reciprocal space
cuts. Such examples will be presented later in this article.
The (hkl) identification of all peaks fitted can be shown by selecting the Show peak label
checkbox (7).
The symmetry of the substrate selected with the combo box (8) can be applied to the fitted
peaks by selecting the checkbox apply substrate symmetry (9)
5.2 Fitting fiber texture on pole figures
To fit a fiber texture, if you have an idea of the plane parallel to the surface, make sure that
(3) is set to zero, select the plane (1), and click on Simul. Fiber (6).
If you don’t know the plane that parallel to the substrate the same procedure as the epitaxial
fitting needs to be apply except the angle (but not 2) can take any value. Once a proper
epitaxial fit has been performed, the user can simulated all the diffraction peaks of the crystal by
setting the delta d value to a larger number such as 1 so all diffraction within ±100% of the dvalue will be considered and plotted on the pole figure when clicking Simul. Epitaxie. The spot
nearest to the center of the pole figure correspond to the fiber texture.
5.3 Fitting axiotaxial texture on pole figures
To fit an axiotaxial texture, you instead of having =0 you need to set and so they
correspond to the center of the axiotaxial texture. If you know the planes that are aligned at the
interface, you just need to use them as selected plane (1), and simulate a fiber using. Simul.
Fiber (6).
If you don’t know the plane that align at the interface, the same procedure as the epitaxial
fitting needs to be apply but a variety of solution will work. Once a proper epitaxial fit has been
performed on the axiotaxial rings, the user can simulated all the diffraction peaks of the crystal
by setting the delta d value to a larger number such as 1 so all diffraction within ±100% of the dvalue will be considered and plotted on the pole figure when clicking Simul. Epitaxie. The spot
nearest to the center of the axiotaxial feature corresponds to the axiotaxial alignment.
6 Producing movie of pole figure
One of the better ways to examine the details of the reciprocal space acquired is to plot it as a
movie of pole figure. In this section we will explain you how to perform this task.
First, the user must select the proper
(1) color min/max
values to be
plotted. To select properly those values
a pole figure must be plot with the
highest resolution possible in d, so no
summation is performed over the pole
figure. In the movie each frame is a
single pole figure with no summations
over d. Therefore delta d needs to be
adjusted to a value of 0.0001 of less
depending on the resolution of your
data in d-spacing, before plotting pole
figures with different color min/max
values.
(3)
Once the color is correctly selected, if
the user does not need to fit anything
on the video he must select or modify a
dummy crystal with too small or too big
lattice spacing (ex: a=9999 b=9999 c
99999) so no diffraction peaks fit in the
d–spacing range selected. For further
details see section 4.3. Otherwise the
user can to fit the proper orientation to
the date has explained previously in
section 5. To simulate fiber texture on
the movie the user push Make movie
fiber (3) and to simulate only epitaxial
components the user select Make
movie (4). The pts. width (9) value will
vary the number of frames around the
optimal value where the diffraction
peaks will be simulated. It is a
percentage of the d-spacing.
(1)
(10)
(11)
(2)
(5)
(6)
(3)
(4)
(9)
(7)
(8)
To fit multiple epitaxial texture on a same pole figure the user needs to find a proper fit and click
add to list (5) to record the values of the fit. Multiple fit can be recorded this way and are
plotted on the video of pole figure when Make movie from list (6) is pushed.
The user has to option to label automatically the peaks on the video by selecting Show peak
label (10) and to apply the substrate symmetry to the fit by selecting Apply substrate symmetry
(11).
The quality of the video can be increase at the cost of space by increasing the quality (7) value
up to 100. The number Frame per second (8) can also be changed.
7 Scanning through reciprocal space
By clicking on Scan applet in the FEP3D window, a new opens with various options.
get coordinate (1): This button allows to get a , coordinate on fig1 (it is using the fig1
value entered in the edit box of the other
applet)
Get 2 coordinates (2): This button allow to
select two (,) coordinates from fig1.
(1)
(27)
(28)
(2)
(29)
(30)
(3)
Get d (3): Allows getting a d-spacing on any (4)
scan of the scan below with d or 2 as x axis.
(5)
Get 2 d or tth (4): Allows getting 2 d-spacing
on any scan of the scan below with d or 2 as
x axis.
tth scan psi phi area (5): Produce a tth scan
summing the intensity over the area defined
with (27) and (28), (29) and (30).
tth scan all psi phi (6): Produce a tth scan
summing over all and .
tth scan specified psi phi (7): Produce a tth
scan at with (27), (29).
(31)
(32)
(33)
(14)
(6)
(7)
(8)
(15)
(16)
(9)
(17)
(10)
(18)
(11)
(12)
(13)
(19)
(20)
(21)
(22)
(23)
(24)
tth scan specified psi (8): Produce a tth scan
at (27) and summing over all .
(25)
(26)
scan selected area (9): produce a scan for
the select area (27) and (28), (29) and
(30) and d-spacing (31), (33).
Area under curve (10): Calculate the area under the tth scan area (5) subtracting a
background defined as line between the curve value at d= (31) and (32). The results appear in
(17)
3D reciprocal space (11): This routine has not been optimized Plot the reciprocal space of a
volume defined by (27), (28), (29), (30), (31), and (32).
7.1 Performing reciprocal space cuts
There are two types of cuts of the reciprocal space that can be performed at this time:
a) Cuts in a plane containing the origin of the reciprocal space and the surface normal
with a selected range of .
To perform those cut the user must first select the range in (27), (28) used (only
values within this range will be considered, we suggest to use 0-90 most of the time)
and the integration range in (29), (30). This range should be tightly chosen near the
peak if the texture is epitaxial or should contain the entire range when fiber or random
texture is present. To perform the cut the user click on vs d intensity graph (12). The
cut will be displayed in different figure depending of the user choice:
(19) plot as reciprocal space uses the normal of the sample as the vertical axis and the
in plane direction as the horizontal axis the results is plot as 1/d so direct vector of the
reciprocal space can be measured.
(20) Real 3D cut plots the reciprocal space within a 3D axes on figure 21. Multiple cuts
will results in multiple planes displayed
(21) sin()^2 plots the intensity as a function of sin()^2 and d. A straight line
correspond to biaxial stress.
(22) sin() plots the intensity as a function of sin() and d.
(23) plots the intensity as a function of and d.
It is possible to perform fit the orientation selected in the main applet on those type of
cut by using the (18) fit vs d button. All the peaks present in the range selected will
be plotted. If the (10) Simul on vs d graph button of the main applet is pushed the
simulation will include all the value possible. This is particularly useful to fit fiber
texture on a vs d graph.
b) Cuts in a plane containing the origin of the reciprocal space and two orientation (, ).
To use this type of cuts two orientation need to be selected using the get coordinate
(24), (25) buttons. Those two orientations with the origin define a plane in reciprocal
space with any orientation possible but always going through the origin. The same
options as in a) exist to plot that plane.
8 Conclusion
In this manual we have presented the main functions of the Fitequipoint program. For further
example we suggest to the reader to look at the video explanation only and the article
submitted to journal of applied crystallography.
i
C. Detavernier, A. S. Özscan, J. Jordan-Sweet, E. A. Stach, J. Tersoff, F. M. Ross, C.
Lavoie, Nature 426, 641 (2003).