pmic12148-sup-0001-SupInfo

Supporting information
Membrane Proteins Isolation and Identification by a
Covalent Binding for Proteome Research
Yiying Liu, Guoquan Yan, Mingxia Gao, Chunhui Deng and Xiangmin Zhang*
Department of Chemistry and Institutes of Biomedical Sciences, Fudan University,
Shanghai, 200433, China
Correspondence: Professor Xiangmin Zhang, Department of Chemistry and
Institutes of Biomedical Sciences, Fudan University, Shanghai, 200433, P. R. China.
E-mail: xmzhang@fudan.edu.cn
Fax: +86-21-6564 1740.
Characterization and measurement of Fe
3
O
4
@PEG-tresyl.
Scanning electron microscopy (SEM) images and energy-dispersive X-ray analysis
(EDX) were obtained at 20 kV on a Philips XL30 electron microscope (Netherlands).
Transmission electron microscopy (TEM) images were acquired at 200 kV from a
JEOL2011 microscope (Japan). Fourier-transform infrared (FT-IR) spectra were collected on a Nicolet Fourier spectrophotometer with KBr wafer as background.
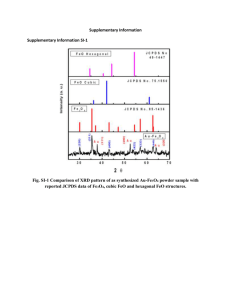
Figure S1A displays the scanning electron microscope (SEM) image of the
Fe3O4@PEG-tresyl, showing a uniform shape and narrow size distribution. Figure
S1B is a transmission electron microscope (TEM) image. The Fe
3
O
4
@PEG-tresyl has a clear core–shell structure with a tresylated PEG shell thickness of about 18 nm. The existence of tresyl group on Fe
3
O
4
@PEG-tresyl was testified by the energy-dispersive
X-ray analysis (EDX) ( Figure S1C ). Both of the S and F elements, which uniquely existed in tresyl group, were detected at a ratio close to 3, indicating the successful immobilization of tresyl group. For further verification, Fe
3
O
4
@PEG and
Fe
3
O
4
@PEG-tresyl were tested by attenuated total reflection Fourier transform infrared (ATR-FTIR) in the range of 3600 cm
-1
~ 300 cm
-1
. In Figure S1D (a) , the peak at 578 cm -1 was from the iron oxide skeleton (–O–Fe), and the peak at 1100 cm -1 originated from the symmetric stretching of the –C–O–C– repeating units on the PEG chains. An absorption band around 3396 cm
-1
should be attributed to the hydroxyl groups at the end of PEG chains, which were anticipated to react with tresyl chloride.
While the corresponding peak of hydroxyl groups in Figure S1D (b) was comparatively flat, and the labeled multiple peaks were assigned to C–F vibration or the sulfonic acid bond. It implied that tresyl group was on the surface of Fe
3
O
4
@PEG.
Fe
3
O
4
@PEG-tresyl nanomaterial exhibits an excellent water dispersibility without any sedimentation.
Figure S1. (A) SEM image of Fe
3
O
4
@PEG-tresyl; (B) TEM image of
Fe
3
O
4
@PEG-tresyl; (C) The energy dispersive X-ray (EDX) spectrum data of the
Fe
3
O
4
@PEG-tresyl material; (D) FTIR spectra of (a) Fe
3
O
4
@PEG, and (b)
Fe
3
O
4
@PEG-tresyl, respectively.
Figure S2. Analysis of membrane proteins capture efficacy by 12 % sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining. Two supernatants (Supernatant 1 and Supernatant 2) were respectively obtained after capturing equal amount of MPs (0.5
g/
L, 240
L) with newly synthesized
Fe
3
O
4
@PEG-tresyl and Fe
3
O
4
@PEG-tresyl stored for one month. The volume of maker is 4 µL, and the volume for each sample is fixed as 10 µL.
Quantification of SDS in the eluate.
The concentration of SDS was measured using a gas chromatograph coupling to a mass spectrometer (GC-MS) according to an experimental strategy.[1] Perkin-Elmer
Clarus 680 gas chromatograph connected to a Clarus SQ8T mass spectrometer
(Waltham, MA, USA) was used. SDS solutions with different concentrations
(0.0003%, 0.001%, 0.003%, 0.006% and 0.03%) were prepared to establish a calibration curve of the peak area of 1-dodecanol. Each solution was 10-fold diluted with 8 mM guanidine-HCl MeOH. One microliters of the solution was injected into the GC-MS system with an injection temperature of 300 °C and in a splitless mode.
The column oven temperature was programmed with an initial temperature of 100 °C for 3 min, and then raised up to 300 °C at a ramping rate of 10 °C/min. The eluted
SDS solutions was detected under the same process and quantified based on the established calibration curve ( see Figure S2 and Table S1 ).
Table S1. Peak areas of SDS with different concentration.
SDS(%) Run 1 Run 2 Run 3
0
0.03
0
89154976
0
81184000
0
82084480
0.006
0.003
0.001
0.0003
21858025
11511961
8616116
2491549
28660938
15735938
8031368
2211481
24559720
12107885
7966573
2856779
Mean
0
84141152
25026228
13118595
8204686
2519936
Figure S3.
Calibration curve for GC-MS measurement of SDS.
Figure S4.
MALDI-TOF MS spectra derived from tryptic digestion peptides of BSA using the CB method without washing (A) and with washing once (B), twice (C), as well as three times (D) by water.
Figure S5. Distribution of identified membrane proteins based on calculated MW (A) and theoretical p I (B).
Reference
[1]Campbell, R.; Winkler, M. A.; Wu, H. Q.
Quantification of sodium dodecyl sulfate in microliter biochemical samples by gas chromatography . Analytical
Biochemistry 2004 , 335 , 98 – 102.