Whole Genome Assembly
advertisement
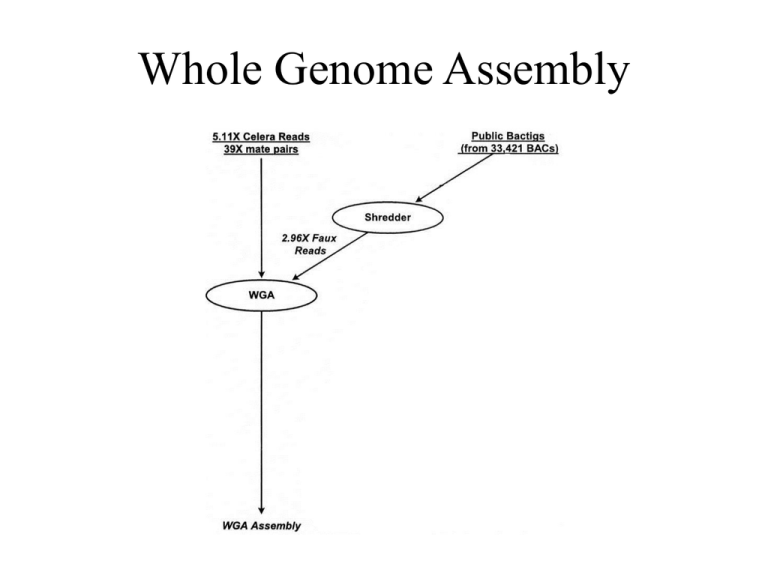
Whole Genome Assembly WGA 1. Screener 2. Overlapper 3. Unitigger, 4. Scaffolder, 5. Repeat Resolver. Overlapper ...looks for end-to end overlaps of at least 40 bp with no more than 6% differences in match. What’s the significance? ...a one in 1017 event. Sequencing Fidelity: 99.96% However ...the Screener doesn’t include all of the “low frequency” level repeats, ...so, a majority of the Overlapper outputs are bogus. Unitigger ...differentiates between a true overlap, and an overlap that includes more than one loci. 8X ...in a world where real data matches expected data, each loci would have 8X coverage, ...over-collapsed. ...if there were repeats, then contigs would be “over-represented”, on average 8 more per repeat. What Now? ... uniquely assembled contigs (unitigs) are readily identifiable, – all of the assembled sequences match over all of the known sequence, - and - ...are consistent with an 8x coverage. Unitigs ...contig cluster is consistent with expected size, ...no dissimilar sequences between any members. ...all other contigs are sent to the Discriminator. Discriminator ...parses the “overcollapsed” contig by using sequence outside of the overlap region Discriminator ...may yield unitigs. Unitigger Output ...correctly assembled contigs covering 73.6% of the genome. Repeat Resolver ...most of the remaining gaps were due to repeats. 1. Allow “low Discriminator Value” contigs to fill gaps, 2. Find BAC sequences that unambiguously match outside the nearest unitig, – 1 in 107 chance of being wrong, 3. Ensure the mate end sequence of candidate BACs match. If that Doesn’t Work ...find a mate-pair that spans the gap, and sequence it, ...make sequencing primer from BES... Chromosome Walking Scaffolder ...contigs the contigs, – uses mate-pair information. WGA Result ...91% sequence, 9% gaps, Compartmentalized Shotgun Assembly Mapping Scaffolds Sequence Tagged Sites STS ...PCR primers are designed for unique regions of the genome or chromosome, ...the chromosome is cut , ...assay two PCR products, frequency of coamplification indicates . Sequence Tagged Sites STS Compartmentalized Shotgun Assembly ...ideally 24, ...really 3845. CSA 92.2 % Sequence 7.8 % Gaps WPA 91 % Sequence 9 % Gaps Chromosome 21 Blue: Gaps Violations: Red : misoriented Yellow: distance PFP CSA Green: Same Order, Orientation Yellow: Same Orientation Red: Out of Order, Orientation Chromosome 8 PFP CSA PFP CSA Major Public Sequence Databases • 281 Curated Data Bases, • “... facilitating Biological Discovery”. What Do We Know? (based on functional group analysis) Science 291 (5507), 1304-1351 Functional Groups 1st GenBank NR protein database was partitioned into clusters using BLASTP, Describing Aligned Sequences 2 nd Statistical descriptions of the cluster are developed and tested, • Hidden-Markov Markers: statistical descriptions of aligned sequences. Functional Group Annotation 3 rd Categorization was done by manual review of the family and subfamily names, ...by examining SwissProt and GenBank records, ...and by review of the literature as well as resources on the World Wide Web. http://www.expasy.ch/cgi-bin/niceprot.pl?P29965 Outcomes? • A relatively small number of structural and functional domains are used in a large number of different proteins, • Pfam: 527 families, • average length is 275 residues, • 456 had “annotated functions”. Nucleic Acids Research 26, 320-322 New Genes 4 th Newly sequenced genes are virtually translated, and the predicted proteins are assayed against raw and HMM databases, ...significance cut-off levels are determined for each functional group family.