Supplementary Table 2. Comparison of assembly outcomes for
advertisement
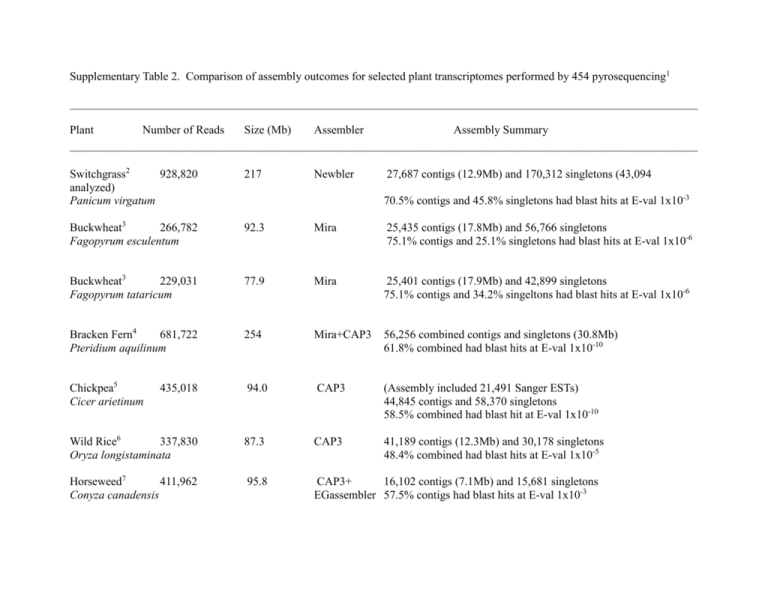
Supplementary Table 2. Comparison of assembly outcomes for selected plant transcriptomes performed by 454 pyrosequencing1 ____________________________________________________________________________________________________________ Plant Number of Reads Size (Mb) Assembler Assembly Summary ____________________________________________________________________________________________________________ Switchgrass2 928,820 analyzed) Panicum virgatum 217 Buckwheat3 266,782 Fagopyrum esculentum 92.3 Mira 25,435 contigs (17.8Mb) and 56,766 singletons 75.1% contigs and 25.1% singletons had blast hits at E-val 1x10-6 Buckwheat3 229,031 Fagopyrum tataricum 77.9 Mira 25,401 contigs (17.9Mb) and 42,899 singletons 75.1% contigs and 34.2% singeltons had blast hits at E-val 1x10-6 Bracken Fern4 681,722 Pteridium aquilinum 254 Mira+CAP3 56,256 combined contigs and singletons (30.8Mb) 61.8% combined had blast hits at E-val 1x10-10 Chickpea5 Cicer arietinum 435,018 94.0 CAP3 (Assembly included 21,491 Sanger ESTs) 44,845 contigs and 58,370 singletons 58.5% combined had blast hit at E-val 1x10-10 Wild Rice6 337,830 Oryza longistaminata 87.3 CAP3 41,189 contigs (12.3Mb) and 30,178 singletons 48.4% combined had blast hits at E-val 1x10-5 Horseweed7 411,962 Conyza canadensis 95.8 CAP3+ 16,102 contigs (7.1Mb) and 15,681 singletons EGassembler 57.5% contigs had blast hits at E-val 1x10-3 Newbler 27,687 contigs (12.9Mb) and 170,312 singletons (43,094 70.5% contigs and 45.8% singletons had blast hits at E-val 1x10-3 Waterhemp8 483,225 Amaranthus tuberculatus 112 CAP3+ 22,035 contigs (9.6Mb) and 22,434 singletons EGassembler 70.7% contigs and 46.0% singletons had blast hits at E-val 1x10-3 Barbados Nut9 Jatropha curcas 13.1 Newbler 17,457 contigs (16.0Mb) and 54,002 singletons 91.2% contigs had blast hits at E-val of 1x10-10 877 Newbler 25,998 contigs (18.0Mb) and 178,363 singletons (trimmed to 381,597 Amaranth10 2,700,168 5,113) Amaranthus hypochondriacus 82.0% isotigs had blast hits at e-value of 1x10-10 ____________________________________________________________________________________________________________ 1 De novo 454 sequencing of plants with limited or no genome sequences. 2 This study. 3 Logacheva MD, Kasianov AS, Vinogradov DV, Samigullin TH, Gelfand MS, Makeev VJ, Penin AA (2011) De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC Genomics 12:30. 4 Der JP, Barker MS, Wickett NJ, dePamphilis CW, Wolf PG (2011) De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genomics 12:99. 5 Hiremath PJ, Farmer A, Cannon SB, Woodward J, Kudapa H, et al. (2011) Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnol J. 2011 May 25. doi: 10.1111/j.1467-7652.2011.00625.x. [Epub ahead of print] 6 Yang H, Hu L, Hurek T, Reinhold-Hurek B (2010) Global characterization of the root transcriptome of a wild species of rice, Oryza longistaminata, by deep sequencing. BMC Genomics 11:705. 7 Peng Y, Abercrombie LL, Yuan JS, Riggins CW, Sammons RD, et al. (2010) Characterization of the horseweed (Conyza canadensis) transcriptome using GS-FLX 454 pyrosequencing and its application for expression analysis of candidate non-target herbicide resistance genes. Pest Manag Sci 66(10): 1053-62. 8 Riggins CW, Peng Y, Stewart CN Jr, Tranel PJ (2010) Characterization of de novo transcriptome for waterhemp (Amaranthus tuberculatus) using GS-FLX 454 pyrosequencing and its application for studies of herbicide target-site genes. Pest Manag Sci 66(10):1042-52. 9 Natarajan P, Parani M (2011) De novo assembly and transcriptome analysis of five major tissues of Jatropha curcas L. using GS FLX titanium platform of 454 pyrosequencing. BMC Genomics 12:191. 10 Délano-Frier JP, Avilés-Arnaut H, Casarrubias-Castillo K, Casique-Arroyo G, Castrillón-Arbeláez PA, et al. (2011) Transcriptomic analysis of grain amaranth (Amaranthus hypochondriacus) using 454 pyrosequencing: comparison with A. tuberculatus, expression profiling in stems and in response to biotic and abiotic stress. BMC Genomics 12:363.