ppt presentation
advertisement

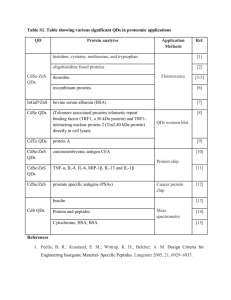
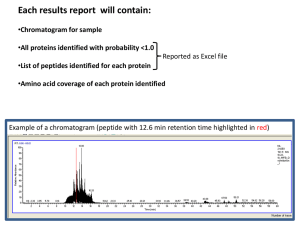
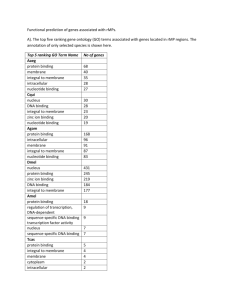
Proteome GENOME Transcriptome Nucleus Proteome Nuclear membrae ER ProteomeMitochondria Golgi Vesicles Chloroplasts Vacuole Set of all protein in certain system - organelles, cells, tissues, organs, organisms, treatments, etc. - includes all protein states (e.g. posttranslational modifications = protein encoded by a single gene is present in many functionally different forms) - e.g. Homo sapiens 30.000 genes, but estimated approx. 500.000 proteins (protein forms) Analysis of protein level and localization Detection of presence of certain protein - by activity determination (possible for some enzymes) - immunodetection: Western blot (after SDS-PAGE), ELISA, … Analysis of certain protein localization - translational fusion with reporter gene (GFP); in situ immunodetection Proteome analysis/comparisons - 2D-electrophoresis - „Gel-free“ methods Detection of selected protein on Western blot Protein electrophoresis (SDS-PAGE) transfer of proteins from gel onto membrane = Western blot Detection by antibodies (primary = interacts with the protein secondary = interacts with primary antibodies from certain species W. blot vizualization usually by enzymatic reaction (color precipitate, chemiluminiscence) Primary antibody Membrane with bound proteins Secondary antibody conjugated with an enzyme or fluorescent label Translational fusion with GFP Removal of stop codone and attachement of GFP gene in reading frame (GFP without stop codone can be even in N-terminus if there is not a localization signal) Translational fusion with GFP Studies of protein localization and protein interactions, Monitoring of different processes and structures in living cells … Golgi apparatus chromozomes microtubules Proteomics - factors influencing proteome Cell cycle phase Interactions with environment Temperature Proteome ??? Stress Physiological state of cell Cell specific gene expression Genome Proteome analysis Before identification, proteins (peptides) has to be fractionated/separated – e.g. 2D-electrophoresis, … Principle of 2-dimensional electrophoresis IEF: isoelectric focusing SDS-PAGE Every spot represents one protein form (amino acid exchange or postranslational modification can shift the position of protein on the gel) First dimension: IEF (isoelectric focusing): - separation of proteins according their isoelectric point (pH with zero charge) - proteins move to the position in gel, where pH matches with their isoelectric point separation of proteins by their charge in pH gradient Second dimension SDS-PAGE - denaturation within the first dimension gel - separation of proteins by size in polyacrylamide gel Principle of 2-dimensional electrophoresis IEF: isoelectric focusing - pH 3 pH 10 200 kD SDS-PAGE - + + 20 kD Staining of protein gels – With silver (nitrate; sensitive, nonquantitative) – Coomassie blue (less sensitive, quantitative) – RI (in vivo labelïng) – Fluorescent dyes (DIGE) - proteome comparisons - difference in gel electrophoresis Protein identification (peptide fingerprinting) • Protein digestion with trypsine or other protease (specific, reproducible cleavage!) • MS (e.g. MALDI/TOF) – Matrix-assisted – Laser desorption / ionisation – Time-of-flight analysis MALDI • Peptides are crystalized in matrix • Laser flash ionizates matrix molecules • Peptides are ionizated with protons transfered from matrix MALDI Matrix-assisted Laser desorption / ionisation • Peptides are crystalized in matrix • Laser flash ionizates matrix molecules • Peptides are ionizated with protons transfered from matrix ToF Time-of-flight (reflects mass) Protein identification by „peptide fingerprinting” Protein digestion (by trypsine) protein I (12 kD) DNA (i EST), protein database 2 fragments (4 kD, 8 kD) protein II (16 kD) 3 fragments (2 kD, 6 kD, 8 kD) generation of theoretical cleavage products MALDI ToF protein I 4000 8000 m/z protein a: protein b: protein c: protein d: protein e: 3, 5, 9, 12 kD 2, 7, 9 kD 4, 8 kD 3, 9, 12 kD 2, 6, 8 kD protein II 2000 6000 8000 m/z Comparison of experimental peptide sizes with the theoretical set generated from proteins in database MS/MS peptide „sequencing“ by MALDI TOF/TOF • Izolated protein or protein mixture is cleaved with protease (alt. LC separation of peptides from protein mixture) • MALDI TOF/TOF – Peptide selection (by TOF) – Peptide fragmentation (collision cell with lower vacuum) – TOF analysis of peptide fragments Mass spectrum of peptide fragments Preferential fragmentation in peptide bond - splitting into two fragments MS spectra analyses iTraq (Isobaric tags for relative and absolute quantitation) „Gel free“ method - an alternative approach to 2D electrophoresis allowing differential labelling and subsequent quantification of proteins from compared samples - protein reproducible cleavage to peptides in each sample - modification of peptides in each sample (chemically same group, but different nonradioactive isotypes) - mixing samples, fractionation - MS/MS analysis - peptide identification - determination of peptide quantity (ratios between samples) iTraq label Ross P L et al. Mol Cell Proteomics 2004;3:1154-1169 - peptide identification (fragmentation spectrum) + - quantification (quantity/ratio of tags) Example of model experiment Modification of amino groups with iTraq reagents 4-(8) x (every sample separately) (every sample with different reagent) Mixing together frakcionace směsi peptidů MALDI TOF/TOF (identification of peptides + ratio of peptides originating from different samples) LC Isoelectric focusing of peptides Protein complex analysis - separation of native complexes (surfice-bound Coomassie BB) - by size in gradient gel Blue Native Gel Electrophoresis SDS-PAGE