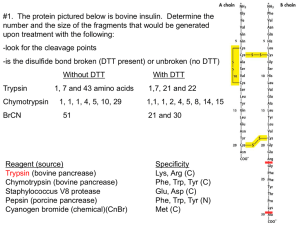
Limited Proteolysis
advertisement

Goals To find the ideal conditions to perform limited proteolysis Most efficient trypsin:AP ratio Buffer solution that optimizes trypsin activity To determine how the reducing agent DTT affects the structural stability of AP Materials and Methods: An Overview First, observe proteolysis of Alkaline Phosphatase by Promega Trypsin Gold Second, trypsinize alkaline phosphatase with and without the reducing agent DTT Third, optimize AP gel electrophoresis conditions Fourth, determine the most efficient ratio of Sigma Trypsin to Alkaline Phosphatase for digestion Proteolysis with Trypsin Gold Take AP, initially at concentration of 1.47 mg/mL, and dilute to 0.5 mg/mL in a 200 mM Tris-HCl buffer, pH 7.4 Bring up lyophilized Trypsin Gold in 200 µL 50 mM acetic acid to a concentration of 0.5 µg/µL 1 Incubate equal parts Trypsin Gold and AP at 37 °C for digestion Extract 15 µL samples at 45 minutes and 90 minutes to monitor digestion This ensured a 1:1 weight ratio of Trypsin:AP Quench digestion for each sample by boiling in water for 2 minutes and subsequently adding 8 µL of loading buffer, boiling again for 5 minutes 2 Run each sample, as well as undigested AP, in a precast 12% acrylamide gel at 200V for ~30 minutes Stain with Coommassie Blue overnight and destain with 10/10 methanol/acetic acid solution Digestion with DTT Using the same procedure as previously, generate trypsin and AP solutions Prepare DTT solution for reduction of AP Add 2 μL DTT stock to 50 μL 0.5 mg/mL AP, making 2 mM DTT in the reaction vessel 3 Also incubate a sample of trypsin for 120 minutes Remove 10 μL samples from each reaction vessel at 5, 15, 30, 45, 60, 90, and 120 minutes Use the same technique as before to quench the digestion Pour two 15% acrylamide gel, one for each reaction condition Incubate the sample in the dark at 50°C for an hour to reduce AP Add 25 μL of 0.5 μg/μL trypsin to the reduced sample and an equal volume of the same concentration unreduced AP and incubate at 37 °C 2 Mix 0.161g DTT into 500μL of H2O to make 50 mM DTT stock solution Load each sample, as well as a molecular ladder, undigested AP, and trypsin, running the gels at 160 V for ~45 minutes Stain and destain as previously described Optimize Electrophoresis Conditions Generate new stock solution of alkaline phosphatase from lyophilized Sigma AP Dilute 6 µL of the stock into 100 µL of 5 mM Tris-HCl, creating a 0.135 µg/µL AP solution Prepare three samples of varying protein concentrations with Loading Buffer to run through SDS-PAGE Bring up in 1mL 5 mM Tris-HCl to a final protein concentration of 4.5 mg/mL 24 µL AP solution with 8 µL Loading Buffer for 3.24 µg AP/Well 18 µL AP solution with 6 µL Loading Buffer for 2.43 µg AP/Well 12 µL AP solution with 4 µL Loading Buffer for 1.62 µg AP/Well Loading Buffer contains 1 part 4X stacking gel buffer, 1.8 part 10% SDS, 0.2 part β-mercaptoethanol, 2 part glycerol, and a dash bromophenol blue Run a precast, 12% resolving gel at 160 volts until AP bands become discernible (20 minutes) Stain and destain as before Ratio Trials of Sigma Trypsin Add 12 µL of 2.5 mg/mL AP to 200 µL 5 mM Tris-HCl, pH 8.0 to create 0.135 µg/µL AP solution 4 Reconstitute lyophilized Sigma Trypsin in 20 µL of 1 M HCl to generate 1 µg/µL Trypsin 5 Dilute Trypsin for 1:2, 1:10, and 1:50 Trypsin:AP ratios by weight for digestion Incubate each sample at 37 °C, removing 10 µL aliquots from each at 5, 30, 60, 105, 150 minutes past initial incubation Trypsin at each ratio should be incubated to monitor any self digestion, only being removed at 150 minutes for analysis Quench digestion for each sample by boiling in water for 2 minutes and subsequently adding 3.3 µL of loading buffer, boiling again for 5 minutes 2 Run one precast 12% (160 V for roughly 40 minutes) for each ratio, with wells for a ladder, each time interval, undigested AP, and Trypsin Stain and destain Day 1 Gel Undigested AP T = 90 min T = 45 min 1:1 Day 3 Gel DTT No DTT Day 4 Gel 1:2 1:50 1:10 Conclusions From the results acquired, a 1:50 trypsin:AP ratio, by weight, yielded the most efficient proteolysis in a 2.5 hour time window Literature research and practical application show a 5mM Tris-HCl at pH 8.0 buffer works for digestion Use low concentrations of AP and Trypsin so that the trypsin can be heavily diluted into Tris-HCl, maintaining a higher pH than the HCl it is brought up in Ideas for Future Research Allow limited proteolysis to take place for a longer period of time, such as 12-24 hours Extract bands with good resolution and analyze samples via mass spectrometry Find which domains are intact after limited proteolysis and Repeat trials with DTT and use other reducing agents known to affect AP Use mass spectrometry to compare the intact domains both with and without reducing agents References 1. 2. 3. 4. 5. Promega Corporation (2009) Technical Bulletin Trypsin Gold, Mass Spectrometry Grade, pp 1-3. Cleveland, D.W., Fischer, S.G., Kirschner, M.W., Laemmli, U.K. (1977) Peptide Mapping by Limited Proteolysis in Sodium Dodecyl Sulfate and Analysis by Gel Electrophoresis. The Journal of Biological Chemistry 242, No. 3, pp 1102-1006. Sigma Aldrich Corporation (2011) Product Information DLDithiothreitol, pp 1. Akitama, Y., Ogura, T., Ito K. (1994) Involvement of FtsH in Protein Assembly into and through the Membrane. I Mutations That Reduce Retention Efficiency of a Cytoplasmic Reporter. The Journal of Biological Chemsitry 269, No. 7, pp 5518-5224. Sigma Aldrich Corporation (2011) Enzyme Explorer Trypsin, pp 1-3.