Symposium Poster - MiddLab
advertisement
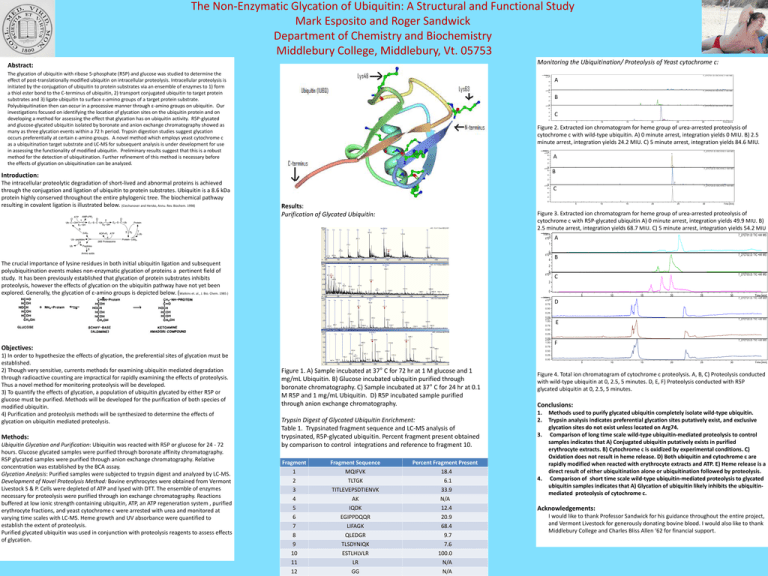
The Non-Enzymatic Glycation of Ubiquitin: A Structural and Functional Study Mark Esposito and Roger Sandwick Department of Chemistry and Biochemistry Middlebury College, Middlebury, Vt. 05753 Monitoring the Ubiquitination/ Proteolysis of Yeast cytochrome c: Abstract: The glycation of ubiquitin with ribose 5-phosphate (R5P) and glucose was studied to determine the effect of post-translationally modified ubiquitin on intracellular proteolysis. Intracellular proteolysis is initiated by the conjugation of ubiquitin to protein substrates via an ensemble of enzymes to 1) form a thiol ester bond to the C-terminus of ubiquitin, 2) transport conjugated ubiquitin to target protein substrates and 3) ligate ubiquitin to surface ε-amino groups of a target protein substrate. Polyubiquitination then can occur in a processive manner through ε-amino groups on ubiquitin. Our investigations focused on identifying the location of glycation sites on the ubiquitin protein and on developing a method for assessing the effect that glycation has on ubiquitin activity. R5P-glycated and glucose-glycated ubiquitin isolated by boronate and anion exchange chromatography showed as many as three glycation events within a 72 h period. Trypsin digestion studies suggest glycation occurs preferentially at certain ε-amino groups. A novel method which employs yeast cytochrome c as a ubiquitination target substrate and LC-MS for subsequent analysis is under development for use in assessing the functionality of modified ubiquitin. Preliminary results suggest that this is a robust method for the detection of ubiquitination. Further refinement of this method is necessary before the effects of glycation on ubiquitination can be analyzed. Intens. x10 6 4 A 2 0 x10 6 6 4 2 7_27CT 02.D: EIC 616.3 +All MS B 0 x10 6 7_27CT 03.D: EIC 616.3 +All MS 6 4 2 C 0 5 10 15 20 25 30 T i me [mi n] Figure 2. Extracted ion chromatogram for heme group of urea-arrested proteolysis of cytochrome c with wild-type ubiquitin. A) 0 minute arrest, integration yields 0 MIU. B) 2.5 minute arrest, integration yields 24.2 MIU. C) 5 minute arrest, integration yields 84.6 MIU. Intens. x10 6 6 4 7_27CT21.D: EIC 616.3 +All MS 1 A 2 0 x10 6 6 Introduction: The intracellular proteolytic degradation of short-lived and abnormal proteins is achieved through the conjugation and ligation of ubiquitin to protein substrates. Ubiquitin is a 8.6 kDa protein highly conserved throughout the entire phylogenic tree. The biochemical pathway resulting in covalent ligation is illustrated below. (Ciechanover and Hersko, Annu. Rev. Biochem. 1998) 7_27CT 01.D: EIC 616.3 +All MS 4 1 7_27CT22.D: EIC 616.3 +All MS B 2 2 0 x10 6 6 C 7_27CT23.D: EIC 616.3 +All MS 1 4 2 0 Results: Purification of Glycated Ubiquitin: 5 10 15 20 25 30 Time [min] Figure 3. Extracted ion chromatogram for heme group of urea-arrested proteolysis of cytochrome c with R5P-glycated ubiquitin A) 0 minute arrest, integration yields 49.9 MIU. B) 2.5 minute arrest, integration yields 68.7 MIU. C) 5 minute arrest, integration yields 54.2 MIU Intens. x109 A 7_27CT01.D: TIC +All MS B 7_27CT02.D: TIC +All MS C 7_27CT03.D: TIC +All MS 1 0 x109 4 The crucial importance of lysine residues in both initial ubiquitin ligation and subsequent polyubiquitination events makes non-enzymatic glycation of proteins a pertinent field of study. It has been previously established that glycation of protein substrates inhibits proteolysis, however the effects of glycation on the ubiquitin pathway have not yet been explored. Generally, the glycation of ε-amino groups is depicted below. (Watkins et. al., J. Bio. Chem. 1985 ) 2 0 x109 2 0 Intens. x10 9 0.75 5 10 15 20 25 30 D Time [min] 7_27CT21.D: TIC +All MS 0.50 0.25 0.009 x10 1.00 0.75 7_27CT22.D: TIC +All MS E 0.50 0.25 0.009 x10 1.00 Objectives: 1) In order to hypothesize the effects of glycation, the preferential sites of glycation must be established. 2) Though very sensitive, currents methods for examining ubiquitin mediated degradation through radioactive counting are impractical for rapidly examining the effects of proteolysis. Thus a novel method for monitoring proteolysis will be developed. 3) To quantify the effects of glycation, a population of ubiquitin glycated by either R5P or glucose must be purified. Methods will be developed for the purification of both species of modified ubiquitin. 4) Purification and proteolysis methods will be synthesized to determine the effects of glycation on ubiquitin mediated proteolysis. Methods: Ubiquitin Glycation and Purification: Ubiquitin was reacted with R5P or glucose for 24 - 72 hours. Glucose glycated samples were purified through boronate affinity chromatography. R5P glycated samples were purified through anion exchange chromatography. Relative concentration was established by the BCA assay. Glycation Analysis: Purified samples were subjected to trypsin digest and analyzed by LC-MS. Development of Novel Proteolysis Method: Bovine erythrocytes were obtained from Vermont Livestock S & P. Cells were depleted of ATP and lysed with DTT. The ensemble of enzymes necessary for proteolysis were purified through ion exchange chromatography. Reactions buffered at low ionic strength containing ubiquitin, ATP, an ATP regeneration system , purified erythrocyte fractions, and yeast cytochrome c were arrested with urea and monitored at varying time scales with LC-MS. Heme growth and UV absorbance were quantified to establish the extent of proteolysis. Purified glycated ubiquitin was used in conjunction with proteolysis reagents to assess effects of glycation. 0.75 7_27CT23.D: TIC +All MS F 0.50 0.25 0.00 5 Figure 1. A) Sample incubated at 37° C for 72 hr at 1 M glucose and 1 mg/mL Ubiquitin. B) Glucose incubated ubiquitin purified through boronate chromatography. C) Sample incubated at 37° C for 24 hr at 0.1 M R5P and 1 mg/mL Ubiquitin. D) R5P incubated sample purified through anion exchange chromatography. Trypsin Digest of Glycated Ubiquitin Enrichment: Table 1. Trypsinated fragment sequence and LC-MS analysis of trypsinated, R5P-glycated ubiquitin. Percent fragment present obtained by comparison to control integrations and reference to fragment 10. Fragment 1 2 3 4 5 6 7 8 9 10 11 12 Fragment Sequence MQIFVK TLTGK TITLEVEPSDTIENVK AK IQDK EGIPPDQQR LIFAGK QLEDGR TLSDYNIQK ESTLHLVLR LR GG Percent Fragment Present 18.4 6.1 33.9 N/A 12.4 20.9 68.4 9.7 7.6 100.0 N/A N/A 10 15 20 25 30 Time [min] Figure 4. Total ion chromatogram of cytochrome c proteolysis. A, B, C) Proteolysis conducted with wild-type ubiquitin at 0, 2.5, 5 minutes. D, E, F) Proteolysis conducted with R5P glycated ubiquitin at 0, 2.5, 5 minutes. Conclusions: 1. 2. 3. 4. Methods used to purify glycated ubiquitin completely isolate wild-type ubiquitin. Trypsin analysis indicates preferential glycation sites putatively exist, and exclusive glycation sites do not exist unless located on Arg74. Comparison of long time scale wild-type ubiquitin-mediated proteolysis to control samples indicates that A) Conjugated ubiquitin putatively exists in purified erythrocyte extracts. B) Cytochrome c is oxidized by experimental conditions. C) Oxidation does not result in heme release. D) Both ubiquitin and cytochrome c are rapidly modified when reacted with erythrocyte extracts and ATP. E) Heme release is a direct result of either ubiquitination alone or ubiquitination followed by proteolysis. Comparison of short time scale wild-type ubiquitin-mediated proteolysis to glycated ubiquitin samples indicates that A) Glycation of ubiquitin likely inhibits the ubiquitinmediated proteolysis of cytochrome c. Acknowledgements: I would like to thank Professor Sandwick for his guidance throughout the entire project, and Vermont Livestock for generously donating bovine blood. I would also like to thank Middlebury College and Charles Bliss Allen ‘62 for financial support.