degradacija proteina
advertisement

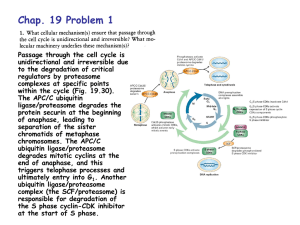
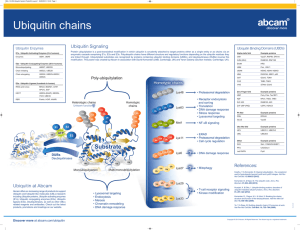
DEGRADACIJA PROTEINA Cartoon depiction of cotranslational folding of a polypeptide. The nascent polypeptide is shown assuming secondary structure as it emerges from the ribosome during the process of biosynthesis. The earliest intermediate, I1, is not wellstabilized by extensive tertiary interactions and is in equilibrium with multiple conformations. The second intermediate shown, I2, is the N-terminal domain; more extensive tertiary interactions will allow this intermediate to be more stable. The final intermediate, I3, depicts the structure of the full-length polypeptide immediately prior to release from the ribosome with the C-terminal domain not yet fully packed. Chaperones and/or folding catalysts (CH and FC) may interact with either the nascent intermediate structures or with the full-length product (M*) following release from the ribosome. The final stages of folding from M* to native monomer, Mn, occur following release. Association of monomeric units into oligomeric structures, O, may occur posttranslationally as depicted or, as discussed in the text, may involve nascent polypeptides. The structure that was used to develop this cartoon was of one subunit of the bacterial luciferase b2 homodimer (66). Suvišne komponente u stanici se proteolitički degradiraju Dva puta degradacije proteina A scenario for GHR endocytosis and downregulation: (1) GH binds and dimerizes two GHRs, which causes recruitment and activation of Jak2; Jak2 phosphorylates the GHR, itself, and STATs. Signal transduction is relayed to the nucleus and genes are activated; (2) specific ubiquitin conjugases (E2) and ligases (E3) dock onto the UbE motif and ubiquitinate the GHR and possibly other attached proteins; (3) the E2/E3s activate the clathrin-coated endocytosis machinery, serving either as adaptors for clathrin or as connectors to AP2 adaptors; (4) transport to early and late endosomes; (5) degradation of GH and GHR by proteasomes and lysosomes. Journal of Cell Science 112, 1417-1423 (1999) Ubiquitination motifs Journal of Cell Science 112, 1417-1423 (1999) Ubikvitin i proteasomi Konjugiranje s ubikvitinom Primjer: Degradacija ciklina tijekom staničnog ciklusa The ubiquitin system of S. cerevisiae The yeast ubiquitin genes, two of which (UBI1 and UBI2) contain introns, encode fusion proteins of ubiquitin (yellow rectangles) to itself (UBI4) or to one of the two specific ribosomal proteins (UBI1–UBI3) (red and blue rectangles). These fusion proteins are cleaved by deubiquitinating enzymes, yielding mature ubiquitin. Thioester bonds between ubiquitin and the active-site Cys residues of ubiquitin-specific enzymes. The conjugation of ubiquitin to other proteins involves a preliminary ATP-dependent step, in which the last residue of ubiquitin (Gly76) is joined, through a thioester bond, to a Cys residue in the ubiquitinactivating (E1) enzyme encoded by UBA1. The activated ubiquitin is transferred to a Cys residue in one of at least 13 distinct ubiquitin-conjugating (E2) enzymes encoded by the UBC family genes, and from there to a Lys residue of an ultimate acceptor protein (yellow oval). This last step and the formation of a multi-ubiquitin chain (black ovals) require participation of another component, called E3 (the names of some of the yeast E3 proteins are included). A targeted, ubiquitinated protein substrate is processively degraded to short peptides by the ATP-dependent 26S proteasome. The N-end rule pathway N-degron Notations in the yeast (a) and mouse (b) pathways show type 1 (purple) and type 2 (red) primary, secondary (light blue) and tertiary (green) destabilizing N-terminal residues; yellow ovals indicate the rest of a protein substrate. a, The in vivo half-lives of X-gals, -galactosidase-based test proteins in S. cerevisiae (right). X-gal proteins bearing stabilizing N-terminal residues (black) are metabolically stable (t1/2, more than 20 h). The tertiary destabilizing residues N (Asn) and Q (Gln) are converted into secondary destabilizing residues D (Asp) and E (Glu) by N-terminal amidohydrolase (Nt-amidase), encoded by NTA1. D and E are conjugated to R (Arg), one of the primary destabilizing residues, by Arg–RNA protein transferase (R-transferase), encoded by ATE1. b, In the mammalian N-end rule pathway, the deamidation step is mediated by two distinct enzymes, NtN-amidase and NtQ-amidase, specific for N-terminal Asn and Gln residues, respectively54. In vertebrates, the set of secondary destabilizing residues contains not only Asp and Glu but also Cys (C), which is a stabilizing residue in yeast55. In mammals but not in yeast, Ala (A), Ser (S) and Thr (T) are primary (type 3) destabilizing residues48. c, S. cerevisiae UBR1 has two binding sites for the primary destabilizing N-terminal residues of either proteins or short peptides. The type 1 site is specific for basic N-terminal residues Arg, Lys and His. The type 2 site is specific for bulky hydrophobic N-terminal residues Phe, Leu, Trp, Tyr and Ile. UBR1 contains yet another substrate-binding site (i), which targets proteins bearing internal (non-N-terminal) degrons. In yeast, these proteins include the CUP9 repressor57. A complex of UBR1 and the ubiquitinconjugating (E2) enzyme RAD6 produces a substratelinked multi-ubiquitin chain Protasomi 20S Proteasome The catalytic core of the proteasome (20S proteasome) is built up of seven different a and seven different bsubunits arranged as a cylindrical 777 7 complex in four stacked rings. Vertebrates possess three additional -subunits that are induced by interferon- (IFN-). Degradation of folded proteins requires cooperation of the 20S-core protease with the 19S cap, a structure that can bind to both ends of the barrel, resulting in the 26S proteasome. The 19S cap contains at least 20 subunits, some of which are involved in the recognition and removal of ubiquitin. It also possesses an ATPase activity that helps to unfold and feed the substrate into a chamber in the 20S core that harbours the active sites. Further complexity arises from an additional 11S regulatory complex, also upregulated upon stimulation with IFN-. Two forms of the 11S regulator exist: a heterooligomeric ring made up of REGa and REGb subunits and a homo-oligomeric REGg particle. The 11S regulator can replace the 19S complex and results in activation and altered peptidase activity upon binding to the 20S proteasome in vitro. Intracellular targeting of proteasome Trends in Cell Biology,10:268, 2000. Multiubiquitinated membrane proteins target the proteasome to the endoplasmic reticulum (ER). Ubiquitin-activating enzyme E1 hydrolyses ATP and forms a high-energy thioester linkage between a cysteine of its active site and the C-terminus of ubiquitin. Activated ubiquitin is then transferred to a ubiquitin-conjugating enzyme E2. These enzymes function in concert with ubiquitin protein ligases, E3, and attach ubiquitin to the substrate protein. Isopeptidases associated with the 19S cap or in the cytosol disassemble the multiubiquitin chain and ubiquitin can be recycled. Proteasomi za antigensku prezentaciju Perfect use of imperfection Nature 404, 709 - 710 (2000) Figure 1 A use for protein waste. A significant amount (30% or more) of protein production is faulty and results in the formation of defective ribosomal products (DRiPs). Cellular quality-control mechanisms ensure that DRiPs are immediately removed by becoming tagged with a small molecule called ubiquitin (Ub) and targeted to the proteasome, which chops the DRiPs up into small peptides. Samples of the peptides are transported by the transporter associated with antigen presentation (TAP) into the endoplasmic reticulum (ER), loaded on MHC class I molecules and transported to the cell surface for recognition by T cells. Peptides from fully functional proteins that have aged are presented to T cells in the same way. But the DRiP short cut ensures that the immune system can get a head start on fighting infection, because it does not have to wait for proteins (such as those produced in virus-infected cells) to age. Dvije glavne obitelji molekularnih chaperona Chaperonin Djelovanje Hsp70 i Hsp60