Natural_Selection
advertisement

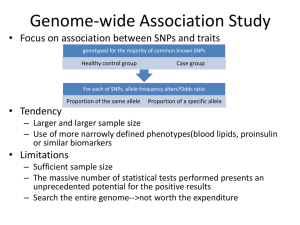
Population genetics, comparative genomics, and natural selection Simon Myers Overview • Identifying selection through – Use of comparative genomic data (FOXP2) – Present day diversity patterns (Lactase) – Both (conserved non-coding regions) Separation of evolutionary timescales • The genome evolves over many millions of years – Our genome almost 99% identical to chimpanzee • Population genetics studies variation among individuals within a population – Uses study of genealogies – In humans, only hundreds of thousands of years • What can population genetics tell us about genome evolution? Targets of selection are important Explain observable phenotypes (Lactase,SLC24A5, EDAR…) Humans Pathogen evolution Disease resistance (LARGE, Duffy) What makes us human? (FOXP2) Other species Resistance to pesticides Adaptive evolution Time • Advantagous mutations arise by chance • Once arisen, carriers have more offspring • “Positive selection” • On average, higher rate of change towards advantageous mutations Looking for positive selection • Direct approach is very difficult – Need to observe trait for long time – Need very strong selection • In many cases, need a more indirect approach – – – – Compare genomes among closely related species Look for “accelerated evolution” Current day patterns of diversity Look for “signature of selection” FOXP2 • Gene coding for a transcription factor • Mutations in this gene cause speech impairment and other problems (Lai et al., Nature 2001) – Mutation in FOXP2 co-segregates with a disorder in a family in which half of the members have severe speech, linguistic and grammatical difficulties – Translocation in same gene in unrelated individual with similar disorder • Are changes in this gene associated with human language development? FOXP2 (Enard et al., Nature 2002) • Are humans different from other species at FOXP2? • Sequence gene in chimpanzee, gorilla, orang-utan, rhesus macaque and mouse • Comparison FOXP2 (Enard et al., Nature 2002) Yellow: human lineage mutations (since chimpanzee-human split) Blue: mutations on all other lineages Very conserved gene (top 5% of 1,880 genes) Only 3 non-repeat amino acid changes in 130 million years between human and mouse 2 occurred on human lineage in last 5-6 million years FOXP2 (Enard et al., Nature 2002) 156 synonymous changes, 0 on human lineage 4 non-synonymous changes 2 on human lineage (p=0.0005 by Fishers exact test) Is this the answer? • Comparative genomics has disadvantages – Need repeated mutations to give power – Tells little about the timescale – Recent research suggests Neanderthals may share FOXP2 mutations with humans (Krause et al., Current Biology 2007) • How do we find out if, and where, we’re currently evolving? Looking for positive selection • Direct approach is typically difficult – Need to observe trait for long time • In many cases, need a more indirect approach – – – – – Compare genomes among closely related species Look for “accelerated evolution” Current day patterns of diversity Look for “signature of selection” Identify effect of selection on diversity patterns Variation data and selection • Revolution in population genetics • Genome-wide datasets – HapMap project – Many unrelated individuals (60 CEU, 60 YRI, 45 JPT and 45 CHB) – Typed at ~4,000,000 loci that vary within population • Allow systematic searches for selection – Comparison of interesting regions to genome – Identification of novel candidates for selection Neutral alleles I II Neutral variation Neutral allele arises III Recombination scrambles variation over time e.g. HapMap The signature of positive selection I II Neutral variation Advantageous allele arises III Spreads (sweeps) rapidly through population Recombination has much less time to scramble variation on selected background The signature of positive selection SelSim (Spencer and Coop, Bioinformatics 2004) The signature of positive selection Neutral mutation at 50% Selected mutation at 50% EHH • Several authors have developed tests based on similar idea – Sabeti et al. (Nature 2002), Voight et al. (PLoS Biology 2006) – Focus on potentially selected mutation – Measure proportion of haplotypes identical, as a function of distance on either side – Compare selected/nonselected types – Look for signal of “extended haplotype homozygosity” (EHH) Simulation results (Voight et al.,PloS Biology 2006) Lactase gene • Most humans lose ability to digest lactose as adults – 70% of all humans are lactose intolerant – In Europe, 95% lactose tolerance Lactase gene • DNA variant C/T-13910 • 14kb upstream of Lactase gene • Completely predicts lactose persistance across human populations (Enattah et al., Nature Genetics 2002) • Mutation enhances promoter activity, so probably causal (Olds et al. Hum. Mol. Genet. 2003) EHH around Lactase From Bersaglieri et al. (AJHG, 2004) EHH around Lactase 5’: p=.012 3’: p<0.0004 Another approach • SNPs that are at highly different frequencies across populations are excellent candidates for selection – SLC24A5 (skin colour, HapMap paper, Lamason et al. Science 2005) – EDAR (hair follicle development, HapMap paper, Sabeti et al. Nature 2007) Testing for ongoing conservation • IDEA: Look at how common variants occurring within CNC’s are in the population – If the CNC’s are functional, mutations in them have a hard time competing – Tend to be rarer in the population than other mutations 42 CNC 5 2 2 4 1 7 5 1 Non-CNC 5 2 3 Purifying selection • Much of the work of selection is removing disadvantageous alleles Maladaptive mutation Fewer offspring Mutation lost • Regions performing some useful function (e.g. genes!) evolve more slowly • Once again, comparative genomics can help! – Look for regions that are conserved between distantly related species Identifying conserved regions 5% of genome is “conserved” – but only 1.5% exonic sequence CNCs • So called conserved non-coding regions (CNCs) make up about 3% of the genome (e.g. Waterson et al. 2002) • Suggests widespread regulatory sequence • Is this stuff real? – Mutational cold spots – Old functionality, now lost • Population genetics enables testing – Approach complements comparative genomics Disadvantageous mutations should be at lower frequency Neutral Negatively selected (2Ns=-2) From talk by S. Williamson SNP frequency “spectrum” in CNC’s • SNPs are at lower frequencies in CNC’s (p=3x10-18) Drake et al. (Nature Genetics, 2005) CNC’s results (Drake et al., 2005) • Shift in frequency spectrum relative to non-conserved regions – Proves conservation is real, and function exists now – Signal robust to demography changes • Signal is comparatively weak! – Not all changes selected against? – Signal stronger nearer genes – Near genes, strength comparable to signal for nonsynonymous mutations in exons • Extreme SNP frequency bias for ultraconserved elements (Katzman et al., Science 2007) – “Ultraconserved elements are ultraselected” Conclusions • Population genetics provides diverse information about molecular evolution • Combining population genetics with knowledge of genomic sequence – New insights into adaptive evolution – Identification of functional sequence • Avalanche of variation data being gathered – Will bring many more insights – Presents major challenges in utilising vast and highly informative datasets, whilst keeping analyses computationally tractable Selected references - Lai, C.S., S.E. Fisher, J.A. Hurst, F. Vargha-Khadem, and A.P. Monaco. 2001. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413: 519-523. - Lamason, R.L., M.A. Mohideen, J.R. Mest, A.C. Wong, H.L. Norton, M.C. Aros, M.J. Jurynec, X. Mao, V.R. Humphreville, J.E. Humbert et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310: 1782-1786. - Olds, L.C. and E. Sibley. 2003. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet 12: 2333-2340. - Sabeti, P.C., D.E. Reich, J.M. Higgins, H.Z. Levine, D.J. Richter, S.F. Schaffner, S.B. Gabriel, J.V. Platko, N.J. Patterson, G.J. McDonald et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832-837. - Sabeti, P.C. P. Varilly B. Fry J. Lohmueller E. Hostetter C. Cotsapas X. Xie E.H. Byrne S.A. McCarroll R. Gaudet et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913-918. - Spencer, C.C. and G. Coop. 2004. SelSim: a program to simulate population genetic data with natural selection and recombination. Bioinformatics 20: 3673-3675. -The International HapMap Consortium. 2005. A haplotype map of the human genome. Nature 437: 1299-1320. - The International HapMap Consortium. 2007. The Phase II HapMap. Nature - Voight, B.F., S. Kudaravalli, X. Wen, and J.K. Pritchard. 2006. A map of recent positive selection in the human genome. PLoS Biol 4: e72. - Waterston, R.H. K. Lindblad-Toh E. Birney J. Rogers J.F. Abril P. Agarwal R. Agarwala R. Ainscough M. Alexandersson P. An et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. Selected references - Bersaglieri, T., P.C. Sabeti, N. Patterson, T. Vanderploeg, S.F. Schaffner, J.A. Drake, M. Rhodes, D.E. Reich, and J.N. Hirschhorn. 2004. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 74: 1111-1120. - Drake, J.A., C. Bird, J. Nemesh, D.J. Thomas, C. Newton-Cheh, A. Reymond, L. Excoffier, H. Attar, S.E. Antonarakis, E.T. Dermitzakis et al. 2006. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet 38: 223-227. - Enard, W., M. Przeworski, S.E. Fisher, C.S. Lai, V. Wiebe, T. Kitano, A.P. Monaco, and S. Paabo. 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418: 869872. - Enattah, N.S., T. Sahi, E. Savilahti, J.D. Terwilliger, L. Peltonen, and I. Jarvela. 2002. Identification of a variant associated with adult-type hypolactasia. Nat Genet 30: 233-237. - Katzman, S., A.D. Kern, G. Bejerano, G. Fewell, L. Fulton, R.K. Wilson, S.R. Salama, and D. Haussler. 2007. Human genome ultraconserved elements are ultraselected. Science 317: 915. - Krause, J., C. Lalueza-Fox, L. Orlando, W. Enard, R.E. Green, H.A. Burbano, J.J. Hublin, C. Hanni, J. Fortea, M. de la Rasilla et al. 2007. The Derived FOXP2 Variant of Modern Humans Was Shared with Neandertals. Curr Biol 17: 1908-1912.