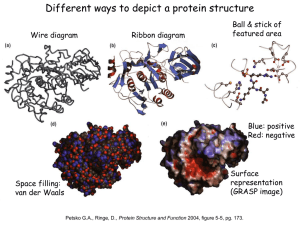
Protein Folding

Proteins
What is a protein?
• A protein is a molecule consisting of amino acids linked in a linear chain through peptide bonds.
Protein primary structure
Peptide formation
There are many kinds of proteins .
• Structural--determine shape and function of cells
• Enzymes--speed up chemical reactions
• Ligand-binding--bind small molecules and transport them to other locations
•
• muscle nerve
Cells
Structural proteins
• collagen -- in connective tissue such as cartilage
• elastin -- in connective tissue such as cartilage
• keratin--in hair and nails
• actin -- in muscle
• myosin -- in muscle to generate mechanical forces
Enzymes
• glucose isomerase--convert glucose into fructose
• rennin--make cheese
• cellulase--break down cellulose into sugars to make ethanol
• amylase--detergent for machine dish washing
Ligand-binding proteins.
• hemoglobin--transport oxygen from the lungs
• antibodies--bind foreign substances for destruction
The string of amino acids tends to “fold” into a shape.
Hemoglobin structure
Heart of Steel (Hemoglobin) by Julian Voss-Andreae
Protein views (Triose phosphate isomerase)
Visualizing proteins
Amino acids
• There are 20 different standard amino acids
• The different amino acids differ in chemical properties.
• Amino Acid
• Alanine
• Arginine
• Asparagine
• Aspartic acid
• Cysteine
• Glutamic acid
• Glutamine
• Glycine
• Histidine
• Isoleucine
• Leucine
• Lysine
• Methionine
• Phenylalanine
• Proline
• Serine
• Threonine
• Tryptophan
• Tyrosine
• Valine
3-Letter 1-Letter Polarity Acidity Hydrophobicity index
Ala A nonpolar neutral 1.8
Glu
Gln
Gly
His
Arg
Asn
Asp
Cys
Ile
Leu
Lys
Met
Phe
Pro
Ser
Thr
Trp
Tyr
Val
I
R
N
D
C
E
Q
G
H
L
K
M
F
P
S
T
W
Y
V polar polar polar nonpolar neutral polar polar polar polar basic (s) -4.5
neutral acidic acidic neutral nonpolar neutral nonpolar neutral neutral neutral nonpolar neutral polar neutral nonpolar neutral
-3.5
-3.5
2.5
-3.5
-3.5
nonpolar neutral polar basic (w)
-0.4
-3.2
nonpolar neutral nonpolar neutral polar basic nonpolar neutral
4.5
3.8
-3.9
1.9
2.8
-1.6
-0.8
-0.7
-0.9
-1.3
4.2
Hydrophobicity index.
• The larger the index, the stronger the tendency to be internal in the protein; the lower the index, the stronger the tendency to appear near the protein surface.
• Amino acids with high index are called hydrophobic ; with low index are called hydrophilic .
What is the shape of the protein?
• This is the “protein folding problem.”
• The geometry and chemistry of the parts of the protein determine how it behaves in the cell.
DNA
• DNA is deoxyribose nucleic acid.
• It occurs as long molecules in a double helix.
DNA is a long molecule in a double helix
What makes DNA?
• DNA consists of sequences of nucleotides.
• There are 4 kinds of nucleotide:
• Adenine (A), Cytosine (C), Guanine (G), and Thymine (T)
Matching
• Each A has weak (“hydrogen”) bonds with T on the other chain.
• Each C has weak (“hydrogen”) bonds with G on the other chain.
A single chain carries the information
• For example, the two strings might be
ACGGTCAG
TGCCAGTC
• Hence all the information is in the order of A, C, G, T in one of the chains.
• We write DNA as a (long) string of A, C,
G, T for example AGGCTACATAG…
Human DNA
• Humans have 46 chromosomes.
• Each chromosome is essentially a double helix of DNA, with variable numbers of nucleotides, from
50,000,000 to 250,000,000 base pairs.
• There are a total of about
2,860,000,000 nucleotide pairs.
Genes
• A gene is a portion of the DNA that tells how to make a protein.
DNA for beta hemoglobin
• ATGGTGCATCTGACTCCTGAGGAGAAGTCTGCCGTTACTG
CCCTGTGGGGCAAGGTGAACGTGGATGAAGTTGGTGGTGA
GGCCCTGGGCAGGCTGCTGGTGGTCTACCCTTGGACCCA
GAGGTTCTTTGAGTCCTTTGGGGATCTGTCCACTCCTGATG
CTGTTATGGGCAACCCTAAGGTGAAGGCTCATGGCAAGAA
AGTGCTCGGTGCCTTTAGTGATGGCCTGGCTCACCTGGAC
AACCTCAAGGGCACCTTTGCCACACTGAGTGAGCTGCACT
GTGACAAGCTGCACGTGGATCCTGAGAACTTCAGGCTCCT
GGGCAACGTGCTGGTCTGTGTGCTGGCCCATCACTTTGGC
AAAGAATTCACCCCACCAGTGCAGGCTGCCTATCAGAAAG
TGGTGGCTGGTGTGGCTAATGCCCTGGCCCACAAGTATCA
CTAA
• Amino Acid
• Alanine
• Arginine
• Asparagine
• Aspartic acid
• Cysteine
• Glutamic acid
• Glutamine
• Glycine
• Histidine
• Isoleucine
• Leucine
• Lysine
• Methionine
• Phenylalanine
• Proline
• Serine
• Threonine
• Tryptophan
• Tyrosine
• Valine
3-Letter 1-Letter Polarity Acidity Hydrophobicity index
Ala A nonpolar neutral 1.8
Glu
Gln
Gly
His
Arg
Asn
Asp
Cys
Ile
Leu
Lys
Met
Phe
Pro
Ser
Thr
Trp
Tyr
Val
I
R
N
D
C
E
Q
G
H
L
K
M
F
P
S
T
W
Y
V polar polar polar nonpolar neutral polar polar polar polar basic (s) -4.5
neutral acidic acidic neutral nonpolar neutral nonpolar neutral neutral neutral nonpolar neutral polar neutral nonpolar neutral
-3.5
-3.5
2.5
-3.5
-3.5
nonpolar neutral polar basic (w)
-0.4
-3.2
nonpolar neutral nonpolar neutral polar basic nonpolar neutral
4.5
3.8
-3.9
1.9
2.8
-1.6
-0.8
-0.7
-0.9
-1.3
4.2
DNA determines the order of amino acids
• ATGGTGCATCTGACTCCTGAGGAGAAGTCTGCCGTTACTG
CCCTGTGGGGCAAGGTGAACGTGGATGAAGTTGGTGGTGA
GGCCCTGGGCAGGCTGCTGGTGGTCTACCCTTGGACCCA
GAGGTTCTTTGAGTCCTTTGGGGATCTGTCCACTCCTGATG
CTGTTATGGGCAACCCTAAGGTGAAGGCTCATGGCAAGAA
AGTGCTCGGTGCCTTTAGTGATGGCCTGGCTCACCTGGAC
AACCTCAAGGGCACCTTTGCCACACTGAGTGAGCTGCACT
GTGACAAGCTGCACGTGGATCCTGAGAACTTCAGGCTCCT
GGGCAACGTGCTGGTCTGTGTGCTGGCCCATCACTTTGGC
AAAGAATTCACCCCACCAGTGCAGGCTGCCTATCAGAAAG
TGGTGGCTGGTGTGGCTAATGCCCTGGCCCACAAGTATCA
CTAA
Primary structure for beta hemoglobin--the order
• MVHLTPEEKSAVTALWGKVNVDEVG
GEALGRLLVVYWTQRFFESFGDLSTP
DAVMGNPKVKAHGKKVLGAFSDGLA
HLDNLKGTFATLSELHCDKLHVDPEN
FRLLGNVLVCVLAHHFGKEFTPPVQA
AYQKVVAGVANALAHKYH
Hemoglobin structure
How does DNA determine the order of amino acids?
• Three successive nucleotides form a
“codon.”
• Different codons stand for different amino acids.
Translating codons
• Ala/A GCT, GCC, GCA, GCG Leu/L
• Arg/R CGT, CGC, CGA, CGG, AGA, AGG Lys/K
• Asn/N AAT, AAC
• Asp/D GAT, GAC
Met/M
Phe/F
• Cys/C TGT, TGC
• Gln/Q CAA, CAG
• Glu/E GAA, GAG
• Gly/G GGT, GGC, GGA, GGG
• His/H CAT, CAC
Pro/P
Ser/S
Thr/T
Trp/W
Tyr/Y
• Ile/I ATT, ATC, ATA
• START ATG
Val/V
STOP
TTA, TTG, CTT, CTC, CTA, CTG
AAA, AAG
ATG
TTT, TTC
CCT, CCC, CCA, CCG
TCT, TCC, TCA, TCG, AGT, AGC
ACT, ACC, ACA, ACG
TGG
TAT, TAC
GTT, GTC, GTA, GTG
TAG, TGA, TAA
DNA for beta hemoglobin
• ATG GTG CAT CTG ACTCCTGAGGAGAAGTCTGCCGTTACTG
CCCTGTGGGGCAAGGTGAACGTGGATGAAGTTGGTGGTGA
GGCCCTGGGCAGGCTGCTGGTGGTCTACCCTTGGACCCA
GAGGTTCTTTGAGTCCTTTGGGGATCTGTCCACTCCTGATG
CTGTTATGGGCAACCCTAAGGTGAAGGCTCATGGCAAGAA
AGTGCTCGGTGCCTTTAGTGATGGCCTGGCTCACCTGGAC
AACCTCAAGGGCACCTTTGCCACACTGAGTGAGCTGCACT
GTGACAAGCTGCACGTGGATCCTGAGAACTTCAGGCTCCT
GGGCAACGTGCTGGTCTGTGTGCTGGCCCATCACTTTGGC
AAAGAATTCACCCCACCAGTGCAGGCTGCCTATCAGAAAG
TGGTGGCTGGTGTGGCTAATGCCCTGGCCCACAAGTATCA
CTAA
Primary structure for beta hemoglobin
• M V H L TPEEKSAVTALWGKVNVDEVG
GEALGRLLVVYWTQRFFESFGDLSTP
DAVMGNPKVKAHGKKVLGAFSDGLA
HLDNLKGTFATLSELHCDKLHVDPEN
FRLLGNVLVCVLAHHFGKEFTPPVQA
AYQKVVAGVANALAHKYH
Hemoglobin structure
The order of amino acids is important
• Consider what may happen when the
“wrong” amino acid is in a certain position.
Primary structure for beta hemoglobin
• MVHLTPEEKSAVTALWGKVNVDEVG
GEALGRLLVVYWTQRFFESFGDLSTP
DAVMGNPKVKAHGKKVLGAFSDGLA
HLDNLKGTFATLSELHCDKLHVDPEN
FRLLGNVLVCVLAHHFGKEFTPPVQA
AYQKVVAGVANALAHKYH
Sickle cell anemia beta hemoglobin
• MVHLTP V EKSAVTALWGKVNVDEVG
GEALGRLLVVYWTQRFFESFGDLSTP
DAVMGNPKVKAHGKKVLGAFSDGLA
HLDNLKGTFATLSELHCDKLHVDPEN
FRLLGNVLVCVLAHHFGKEFTPPVQA
AYQKVVAGVANALAHKYH
• Amino Acid
• Alanine
• Arginine
• Asparagine
• Aspartic acid
• Cysteine
• Glutamic acid
• Glutamine
• Glycine
• Histidine
• Isoleucine
• Leucine
• Lysine
• Methionine
• Phenylalanine
• Proline
• Serine
• Threonine
• Tryptophan
• Tyrosine
• Valine
3-Letter 1-Letter Polarity Acidity Hydrophobicity index
Ala A nonpolar neutral 1.8
Glu
Gln
Gly
His
Arg
Asn
Asp
Cys
Ile
Leu
Lys
Met
Phe
Pro
Ser
Thr
Trp
Tyr
Val
I
R
N
D
C
E
Q
G
H
L
K
M
F
P
S
T
W
Y
V polar polar polar nonpolar neutral polar polar polar polar basic (s) -4.5
neutral acidic acidic neutral nonpolar neutral nonpolar neutral neutral neutral nonpolar neutral polar neutral nonpolar neutral
-3.5
-3.5
2.5
-3.5
-3.5
nonpolar neutral polar basic (w)
-0.4
-3.2
nonpolar neutral nonpolar neutral polar basic nonpolar neutral
4.5
3.8
-3.9
1.9
2.8
-1.6
-0.8
-0.7
-0.9
-1.3
4.2
Simple model
• Pretend there are only 2 kinds of amino acid--H and P.
• H stands for “hydrophobic”.
• Pretend that they must be placed on a grid.
• Example: HHPPPPPPPHH
A folding of HHPPPPPPPHH
H
H
H
H P
P
P
P
P
P
P
Another folding of
HHPPPPPPPHH
H
H
P P
H
H
P
P
P P
P
Energy
• HH has energy -1.
• PP has energy 0.
• HP has energy 0.
• PH has energy 0.
• The protein folds so as to minimize the energy.
A folding of HHPPPPPPPHH with energy -2
H
H
P
P
P
P
P
H
H P P
A folding of HHPPPPPPPHH with energy -4
H
H
P P
H
H
P
P
P P
P
A folding of HHPPPPPPPHH with ? energy
H
H
H
H
P
P
P
P
P P
P
The real problem
• There are 20 amino acids.
• Pairs have different energies.
• Typically a protein has about 100 amino acids.
• The protein is in 3 dimensions.
• It does not need to be on a grid.
• It must be worked on a computer.
The Direct Approach
• Write down a formula for the energy E, taking into account the (variable) locations of all amino acids, all charges and electrostatic attractions and repulsions, and all constraints.
• Minimize E.
Indirect Methods
• Statistics of amino acids in known structures
• Neural network models
• Nearest neighbor methods
• Hidden Markov models
Does a method work?
• We want to be able to check some answers, to see whether a method appears to work.
• Professor Zhijun Wu works on some problems related to this.
NMR
• NMR is Nuclear Magnetic Resonance
• Using NMR one can often find the distances between some particular atoms in a protein.
A1 d(1,4)
A4
Distances
A2 d(2,3)
• Here d(1,4) is the distance between the first and fourth atoms.
A3
A1 d(1,4)
Locations
A2
• A1 is at (x11, x12, x13).
• A2 is at (x21, x22, x23).
• A3 is at (x31, x32, x33).
• A4 is at (x41, x42, x43).
d(2,3)
A3
• Once you know all the locations, you know the shape of the protein.
A4
Position Matrix
• Form the matrix X
A1 d(1,4)
A2 d(2,3)
A3
A4 x11 x12 x13 x21 x22 x23 x31 x32 x33 x41 x42 x43
A1 d(1,4)
Matrix Equation
• It turns out that
A2 d(2,3)
A3
A4
X X T = D where D is a matrix that can be obtained just using all the numbers d(i,j).
A1 d(1,4)
The matrix D
A2 d(2,3)
A3
• If there are n atoms and the last is at the origin, then the entry of D in the ith row and jth column is
(d(i,n) 2 - d(i,j) 2 + d(j,n) 2 ) / 2
A4
Solving the matrix equation
A1
A2 d(2,3)
• Professor Zhijun Wu studies ways to solve such matrix equations rapidly.
A3 d(1,4)
A4
Energy
• HH has energy -1.
• PP has energy 0.
• HP has energy 0.
• PH has energy 0.
• The protein folds so as to minimize the energy.
What is the best folding of
• HPPHPPHPHPPHPHPHHH
• (Careful: answer is on the next slide)
HPPHPPHPHPPHPHPHHH
P
H
P H
P
H
H
P
H
H
P
H
H
P
P
H P
P
with energy -11