Metabol Nutri-ClinEnz Med 2_6 Nov 2012
advertisement

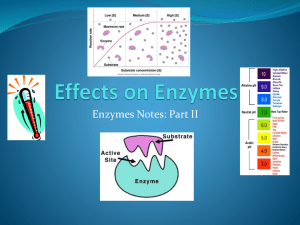
Metabolism & Nutrition 2012 Clinical Enzymology (Prof. Dr. Jerapan Krungkrai) Objectives & Contents: • Clinical correlations of enzymes • Enzymes as markers for diagnosis • Enzymes used in therapy • Enzymes as target of drug • Enzyme abnormalities in metabolisms Enzymes: properties and measurements (I) Nature: catalytic proteins / denaturation property enzyme Substrate Product cofactor/coenzyme 3-dimensional structure: monomer oligomer Variants:1) isozymes (different genes)- tissue-specific forms 2) allozymes (different alleles at single genetic locus) 3) post-translational modifications- cell and tissue specific forms, E.g., liver- and bone-specific alkaline phosphatase (ALP) differ only in carbohydrate contents attached to the ALP proteins. Enzymes: properties and measurements (II) Measurements: 1) kinetic properties and activity of enzyme [S], [P], Km, v, Vmax 2) expression of activity measurements 1 unit of enzyme activity (U) = 1 micromol per min. Then, U/l of sample or U/ml of sample (e.g.,plasma, serum, blood, urine etc.) are widely used. 3) direct measurement of enzyme protein concentration. This is limited due to very low concentrations of proteins. I. Clinical Correlations of Enzymes: Principles of Diagnostic Enzymology 1. Introduction: A human cell contains 28,602 different proteins, 2,709 proteins of which are enzymes. They are assigned roles in ~ 135 metabolic pathways (2,645 metabolites) They are distributed in: cytosol, nucleus, rough and smooth ER, Golgi bodies, mitochondria, lysosomes, plasma and organellar membranes. The enzymes are mainly synthesized in cytosol or ER (stay, or target to other organelles and export to extracellular environment, or extracellular enzymes). A relatively small numbers are synthesized in the mitochondria and these enzymes stay within this space. The activity of an intracellular enzyme is determined by the rates of synthesis, inactivation and degradation (= turnover). Figure 1. Turnover of intracellular enzymes Nobel Prize Chemistry 2004: Rose, Hershko, Ciechanover Figure 2. Mechanism of enzyme release from damaged cell Table 1. Half-lives of clinically important enzymes in plasma Enzyme Range (hours) Lactate dehydrogenase (LD) LD-1 (H4) 50-70 LD-5 (M4) 8-14 Alanine transaminase (ALT, GPT) 40-50 Aspartate transaminase (AST, GOT) mitochondrial AST 6-7 cytosolic AST 12-17 Creatine kinase (CK) CK-MM 10-20 CK-MB 7-17 CK-BB 3 Alkaline phosphatase (ALP) liver ALP 190-230 bone ALP 30-50 Most enzymes are present in cells at much higher concentrations than in plasma. Some occur predominantly in cells of certain tissues. Normal plasma enzyme level (normal or reference range, e.g., 5’-nucleotidase, ALP, amylase =2-15, 30-95, 95-290 U/L). The normal levels reflect the balance between the rate of synthesis and release into plasma during cell turnover, and the rate of clearance from the circulation. The enzyme level in plasma may be: •increased due to proliferation of cells, an increase in rate of cell turnover or damage or in enzyme synthesis, or to reduced clearance from plasma; •lower than normal, due to reduced synthesis, congenital deficiency. Table 2. Serum normal (reference) ranges of clinical enzymes Enzyme Abbreviation Range (male> female) Acid phosphatase ACP, AP Alkaline phosphatase ALP Alanine transaminase ALT, G PT Aspartate transaminase AST, GOT Alpha-amylase AMS Aldolase ALS Creatine kinase CK, CPK Gamma-glutamyl transferase GGT Glucose-6-phosphate dehydrogenase G-6-PD Lactate dehydrogenase LD, LDH Lipase LPS Leucine aminopeptidase LAP 5’-Nucleotidase 5’NT Pseudocholineesterase PChE Ceruloplasmin (Copper-oxidase) 0.2-5.0 U/L 30-95 U/L 6-37 U/L 5-30 U/L 95-290 U/L 1.5-8.0 U/L 15-160 U/L 6-45 U/L 0-0.2 U/L 100-225 U/L 0-2 U/ml 11-30 U/L 2-15 U/L 5-12 U/ml 0.2-0.6 g/L Stability + +++ ++++ +++ ++++ ++++ -++++ +++ + ++++ +++ +++ ++++ 2. Assessment of cell damage and proliferation: •Changes in plasma enzyme levels may help to detect and localize tissue cell damage or proliferation, or to monitor treatment and progress of disease. •Plasma enzyme levels depend on: a) the rate of release from damaged cells which, in turn, depends on the rate at which damage is occurring; b) the extent of cell damage. •In the absence of cell damage, the rate of release depends on: a) the rate of cell proliferation; b) the degree of induction of enzyme synthesis. •These factors are balanced by : a) the rate of enzyme clearance from the circulation (only partly known); b) half-life of enzyme. II. Enzymes as markers for diagnosis Introduction: Isozymes are enzymes catalyzing the same catalytic reaction, but they are synthesized from different genes/loci, and most contain subunits (quaternary structure). This will result in cell and tissue-specific forms of isozymes. Examples of some clinically significant isozymes are as follows: 1. LDH M4 M3H M2H2 MH3 (muscle) LD-5 LD-4 LD-3 LD-2 - ve ------- electrophoresis -------> H4 subunit LD-1 (heart) + ve 2. ALP kidney (isozyme I) <intestine <placenta, kidney (isozyme II) <bone <liver -ve --------- electrophoresis --------> +ve 3. CK CK-3, MM (heart, muscle) CK-2, MB (heart >> muscle) CK-1, BB (brain) -ve --------- electrophoresis --------> +ve 4. ACP Prostate ACP >>> liver, red cells & bone ACPs - ve L-tartrate (prostatic acid phosphatase, PAP) 5. Prostatic specific antigen (PSA), a serine protease (chymotrypsin type), functions in clot liquefaction of semen coagulation. Table 3. Enzyme markers of clinical significance Enzyme (abbreviation) Clinical significance Acid phosphatase (ACP) Prostatic carcinoma Alkaline phosphatase (ALP) Obstructive liver diseases, bone disorders Alanine transaminase (ALT,GPT) Hepatic disorders, viral hepatitis Aspartate transaminase (AST,GOT) Myocardial infarction, hepatic disorders Alpha-amylase (AMS) Acute pancreatitis Aldolase (ALS) Skeletal muscle disorders Creatine kinase (CK) Myocardial infarction, muscle disorders Gamma-glutamyl transferase (GGT) Hepatic disorders G-6-PD Drug-induced hemolytic anemia Lactate dehydrogenase (LD) Myocardial infarction, hepatic disorders, carcionomas Lipase (LPS) Acute pancreatitis Leucine aminopeptidase (LAP) Hepatobiliary disorders 5’-Nucleotidase (5’NT) Hepatobiliary disorders Pseudocholineesterase (PChE) (butyrylcholine as substrate) Ceruloplasmin (Copper-oxidase) Organophosphate poisoning Wilson’s disease (abnormal Cu metabolism) Plasma enzyme patterns in disease: diagnosis & monitor Table 4. Time sequence of changes in plasma enzymes after myocardial infarction (hours, h; days, d) Enzyme Onset of Peak activity Degree of elevation (h) (h) elevation Duration of elevation (d) CK (total) 4-8 12-24 5-10 x normal 3-4 CK-MB 4-8 24-36 5-15 x normal 2-3 8-12 24-48 2-3 x normal 4-6 LD 12-24 48-72 2-3 x normal 10 LD-1>LD-2 12-24 AST (GOT) Remarks: Precision of diagnosis can be improved by • estimations of more than one enzyme • isozyme determinations • serial enzyme estimations 5 Figure 3. Time course of enzymes release into plasma during infarction III. Enzymes as therapeutic agents, drugs Table 5. Enzymes used in therapy are genetically engineered proteins. Enzyme Disease/therapy Protease, e.g., Streptokinase, Clot lysis in myocardial Activase(plasminogen activator) infarction, trauma, bleedings Aspariginase, Acute lymphocytic leukemia e g., Oncospar (pepasparagase) Adenosine deaminase, e.g., Adagen Superoxide dismutase, Severe combined immunodeficiency syndrome (SCID) Head injury (clinical trial phase) e.g., Dismutec peg Nanoenzyme/nanozyme (2007)-catalase Parkinson’s disease(attenuate neuroinflammatory process) IV. Enzymes as drug targets Table 6. Enzyme targets for drugs in clinical use. Enzyme targeting Drug Dihydrofolate reductase Antifolates: methrotrexate (cancer), (Folate metabolism) pyrimethamine (protozoa, malaria) trimethoprim (bacteria) Xanthine oxidase Allopurinol (hyperuricemia, gout) (Purine metabolism) Thymidylate synthase 5-Fluorouracil & (Pyrimidine metabolism) 5-fluorodeoxyuridine (cancer) Glycopeptide transpeptidase Antibiotics, penicillin HIV-Reverse transcriptase 3’-azido-2’,3’-dideoxythymidine (AZT) HIV & SARS proteases Ritonavir, saquinavir (clinical trial phase) Rational drug design, based of known targeting enzyme A). Acquired immunodeficiency syndrome (AIDS) & Bird Flu A.1. HIV-1 Protease: HIV-1 has an aspartate protease, which is essential for delivery of structural and functional processing of the gag and gag-pol viral gene products. Figure 4. Protease inhibtor design, with known 3-D structure at active site complexed with the drug with a Ki of 0.25 nM A.2. HIV-1 Reverse Transcriptase (RT): RT is a viral polymerase responsible for synthesis of viral DNA strand. The RT inhibitors are majority for drugs in clinical use. K103L mutation -> HIV-1 resist to drugs1-3 Figure 5. Crystal of drug 4 A.3. Avian flu, H5N1 virus: Neuraminidase (NA) Enzyme NA is responsible for release of virion from the host epithelial cells by cleaving the receptor sialic acid. NA inhibitor is the drug in clinical use, e.g., zanamivir (Relenza) and oseltamivir (Tamiflu) Tamiflu Figure 6. Rational design of Tamiflu on viral NA Crystal of NATamiflu complex B). Alzheimer’s disease (AD) B.1. AD involves mutation of gene APP (chromosome 21) coding for amyloid beta protein, this APP deposits in neurons of brain as amyloid fibrils -->and leads to cell damage. Loss of cholinergic cell is accompanied by reduced : - ChAT(choline acetyltransferase), ACh (acetylcholine), - AChE (acetylcholine estease). This is compensated by increased BuChE (butyrylcholinesterase) from Glial cell. Figure 7. -ve -ve Rational design is to inhibit the BuChE, ACh is not destroyed. Fig 8. Abnormal or mutant APP (amyloid beta protein) deposits in neurons as amyloid fibrils --> cell damage. B.2. Design of drug for treatment of Alzheimer’s disease : Cholinesterase as enzyme target, phenothiazine derivatives as drug. Figure 9. V.Enzymes abnormalities in metabolisms 1. Excess enzyme activity Gout is characterized by elevated uric acid levels in blood and urine, due to overproduction of de novo purine nucleotides. E.g., Excess PRPP synthase activity (Xlinked recessive inheritance pattern) + ve [PRPP] purine nucleotides then leads to increase degradation of purines to uric acid through xanthine oxidase. Figure 10. 2. Enzyme deficiency Identification and treatment of enzyme deficiency. Enzyme deficiencies usually lead to increased accumulation of specific metabolites in plasma and hence in urine. This is useful in pinpointing enzyme defects. E.g., De novo pyrimidine pathway: defects of OPRT and OMPDC leads to accumulation of orotate ----> Hereditary orotic aciduria (Gene mapping, 3q13; inheritance pattern, autosomal recessive). Figure 11. Uridine 3. Enzyme defects found in all human metabolisms. Table 7. Examples of enzyme defects. Enzyme defect Disease Metabolism/molecule involved Pyruvate kinase Deficiency/Anemia Glycolysis Pyruvate dehydrogenase Pyruvate/Krebs cycle Chronic lactic acidosis G-6-PD Deficiency Pentose phosphate pathway Glycogen debranching enzyme Cori (type III ) Gylcogen storage Iduronate sulfatase Hunter Mucopolysaccharides Acyl-CoA dehydrogenase Deficiency Fatty acid oxidation Hexoaminidase A Tay-sachs Lipid/sphingolipid storage Acid lipase Deficiency Cholesterol/Triacylglycerol (TAG) HGPRT Lesch-Nyhan Purine OPRT/OMPDC Orotic aciduria Pyrimidine Phenylalanine hydroxylase Amino acids/Phe Phenylketonuria Arginase Deficiency Amino acids/Arg /Urea cycle Lysyl hydroxylase Ehlers-Danlos Collagen List of References: • Moss, D.W., and Rosalki, S.B. (1996) Enzyme Test in Diagnosis, Arnold Group, London. • Mayne, P.H. (1996) Clinical Chemistry in Diagnosis and Treatment, 6th edition, Arnold Group, London. • Cohn, R.M., and Roth, K.S. (1996) Biochemistry and Diseases, chapters: 11, 12, 15-17 & 25, Williams& Wilkins, Baltimore. • Devlin, T.M. (2002) Textbook of Biochemistry with Clinical Correlations, pp. 166-174, 5th edition, Wiley & Sons, New York. • Murray, R.K., et al. (2002) Harper's Biochemistry, pp. 8183, 26th edition, Appleton & Lange, Stamford. Evaluation: MCQs, 4 questions/5 choices Good Luck Role of telomerase in telomere replication in eukaryotes Shampay J, Szostak JW, Blackburn EH in Nature. 1984 Jul 12-18;310(5973):154-157. DNA sequences of telomeres maintained in yeast. Role of telomerase in telomere replication in eukaryotes 1. Enzyme telomerase activity is repressed in somatic cells of multi-cellular organisms, resulting in chromosome shortening with each cell generation. This may be important in cell aging. 2. In many cancer cells, there is reactivated telomerase activity. 3. DE Shippen (1990) found that older (70-year old) human chromosomes are shorter than from 20 years old ones, due to reduced telomerase activity. 4. Recently, the shortening of telomere is associated with atherosclerosis and diabetes mellitus type II. [Ref: KD Salpea & SE Humphries (2010), Atherosclerosis 209, 35-38]