Practice
advertisement
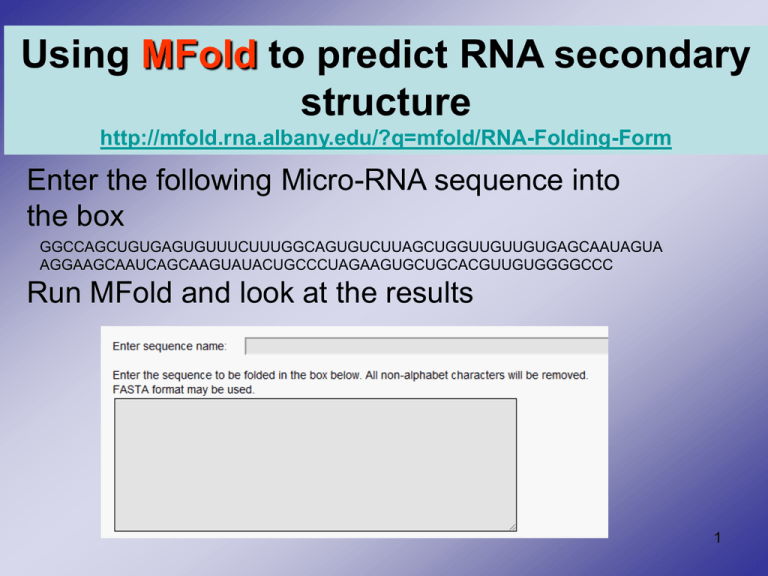
Using MFold to predict RNA secondary structure http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form Enter the following Micro-RNA sequence into the box GGCCAGCUGUGAGUGUUUCUUUGGCAGUGUCUUAGCUGGUUGUUGUGAGCAAUAGUA AGGAAGCAAUCAGCAAGUAUACUGCCCUAGAAGUGCUGCACGUUGUGGGGCCC Run MFold and look at the results 1 Constraint information • Force bases 30-35 to be single stranded • Is the result different? 2 PSIPRED (Protein Structure Prediction Server) Secondary Structure Prediction http://bioinf.cs.ucl.ac.uk/psipred/ 1. Use the protein NP_360043. 2. Paste the sequence without the header line. 3. Enter your UH email address. 4. Click the Predict button. 5. View the results. 3 Helix Coil Beta Strand 4 Helix Beta Strand Coil 5 3-D structures Retrieving and displaying a 3-D structure from PDB http://www.rcsb.org/pdb/ 1. Enter the protein's name and click search 2. View the protein with KiNG viewer 3. Examine the information and links to other databases 8 Protein visualization with FirstGlance in Jmol http://molvis.sdsc.edu/fgij/ Secondary Structure Alpha Helices Beta strands Random coils 10 MMDB • NCBI's structure database is called MMDB (Molecular Modeling DataBase), and it is a subset of experimentally derived threedimensional structures obtained from the Protein Data Bank (PDB) (excluding theoretical predictions) • http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.shtml VAST • NCBI creates and maintains a database of structure alignments, called VAST, for all pairs of proteins from MMDB whose structures have some similar core regions. They are called “Structural neighbors”. • http://www.ncbi.nlm.nih.gov/Structure/VAST/vast.shtml Example • Type "1D5R" in the query box and hit "Get." This brings up the MMDB summary page. 14 • The graphic is saying that the protein is composed of a single chain (A) that NCBI has parsed into two domains: the N-terminal domain 1, and C-terminal 2. Clicking on the top bar marked "Chain A" will bring up VAST neighbors based on the whole chain, while clicking on the colored regions marked "1" or "2" will show neighbors of these individual domains. 16 Click on the “1” domain to get proteins that consists of a similar structure like the PTPc domain 17 Lets view the "3D Alignment” of 1D5R:A with 2BZL:A using the CE tool http://cl.sdsc.edu/ce.html Crystal Structure Of The Human Protein Tyrosine Phosphatase N14 At 1.65 A Resolution 19 20 The two sequences have a 20.7% sequence similarity. The root mean square deviation (RMSD) in terms of structural distance is 2.47. Now lets compare hemoglobin A (4HHB:A) and B (4HHB:B)? 21 Hemoglobin A and B have a sequence similarity of 40.3 % and a root mean square deviation of 1.49. 22 Tertiary structure prediction • From ExPASy http://www.expasy.org/tools/, you can find links to many prediction servers: Tertiary structure prediction • For example the HMMSTR/Rosetta server predicts protein structure from sequence (Ab initio). Protein 3D structure prediction HMMSTR/Rosetta Web Server