Conditional Random Field(CRF)
advertisement

Presenter: Kuang-Jui Hsu
Date :2011/6/16(Thu.)
Outline
1. Introduction
2. Graphical Models
2-1 Definitions
2-2 Application of Graphical Models
2-3 Discriminative and Generative Models
3. Linear-Chain Conditional Random Fields
3-1 From HMMs to CRFs
3-2 Parameter Estimation
3-3 Inference
Introduction
Relational data:
Statistical dependencies exist between the entities
Each entity often has rich set of features that can aid classification
Graphical models are a natural formalism for exploiting the
dependence structure among the entities
We can use the two different types of models to model it
Markov random field (MRF)
Conditional random field (CRF)
Two Types of Models
MRF
The traditional type of model
Modeled by the joint distribution 𝑝 𝒚, 𝒙 ,where 𝒚: the data we want
to predict, and 𝒙: the data we can observe or called features
It requires modeling 𝑝 𝒙 , but 𝑝 𝒙 is hard to be modeled when
using rich features which often include the complex dependences
Ignoring the features can lead to reducing the performance
CRF
Modeled by the conditional distribution 𝑝 𝒚|𝒙
Specifies the probabilities of label data given observation data, so
not to model the distribution 𝑝 𝒙 even with rich features
Solve the problem using the MRF
Introduction
Divided into two part
Present a tutorial on current training and inference technique for
CRF
Discuss the important special case of linear-chain CRFs
Generalize these to arbitrary graphical structure
Present an example of applying a general CRF to a practical
relational learning problem
Outline
2. Graphical Models
2-1 Definitions
2-2 Application of Graphical Models
2-3 Discriminative and Generative Models
Definitions
Probability distribution over sets of random variables:
𝑉 =𝑋∪𝑌
𝑋: a set of input variables we assumed are observed
𝑌: a set of output variables we wish to predict
An assignment to 𝑋 by x and an assignment to a set 𝐴 ⊂ 𝑋
by 𝒙𝐴 , and similar for Y
𝟏 𝑥=𝑥 ′
1, 𝑥 = 𝑥 ′
=
0, otheriwise
Definitions
A graphical model is a family of probability distributions
that factorize according to an underlying graph.
There are two type of graphical models
Undirected graphical models by using the factors to represent
Directed graphical models by using the Bayesian concept
Undirected Graphical Model
Given a collection of subsets 𝐴 ⊂ 𝑉, define an undirected
graphical model form:
1
𝑝 𝒙, 𝒚 =
𝑍
Ψ𝐴 (𝒙𝐴 , 𝒚A )
𝐴
For any choice of factors 𝐹 = {Ψ𝐴 }, where Ψ𝐴 : 𝒱 𝑛 → ℛ +
Z: a normalization factor defined or called a partition
function as
𝑍=
Ψ𝐴 (𝒙𝐴 , 𝒚A )
𝒙,𝒚
𝐴
Computing the Z is a difficult work, but much work exists on
how to approximate it
Undirected graphical models
Present the factorization by a factor graph
1
𝑝 𝒙, 𝒚 =
𝑍
Ψ𝐴 (𝒙𝐴 , 𝒚A )
𝐴
A bipartite graph G=(V, F, E)
Factor nodeΨ𝐴 ∈ 𝐹
Variable node 𝑣𝑠 ∈ 𝑉
Undirected graphical models
Assume that each Ψ function has the form:
Ψ𝐴 𝒙𝐴 , 𝒚A = exp(
𝜃𝐴𝑘 𝑓𝐴𝑘 (𝒙𝐴 , 𝒚𝐴 ))
𝑘
For some real-valued parameter 𝜃𝐴 , and for some set of feature
function {𝑓𝐴𝑘 }
Directed graphical models
Also known as a Bayesian network
𝑝 𝒚, 𝒙 =
𝑃(𝑣|𝜋(𝑣))
Based on a directed graph 𝐺 = 𝑉, 𝐸
𝑣∈𝑉
Factorized as:
The parent of the variable 𝑣
𝑝 𝒚, 𝒙 =
𝑃(𝑣|𝜋(𝑣))
𝑣∈𝑉
𝜋(𝑣): the parent of the variable 𝑣
The variable 𝑣
Generative model
Generative model: call directed graphical models
satisfied the condition that no 𝑥 ∈ 𝑋 can be a parent of an
output y ∈ 𝑌
A generative model is one that directly describes how the
outputs probabilistically “generate ” the inputs
Outline
2. Graphical Models
2-1 Definitions
2-2 Application of Graphical Models
2-3 Discriminative and Generative Models
Application of Graphical Models
Devote special attention to the hidden Markov
model(HMM), because of close relation to the linear-
chain CRF
Classification
Introduce two type of the classifier
Naïve Bayes classifier: based on a joint probability model
Logistic regression: based on a conditional probability model
Naïve Bayes Classifer
Predicting a single class variable y given a vector of features
𝐱 = 𝑥1 , 𝑥2 , … , 𝑥𝑘
Assume that all the features are independent
The resulting classifier is called naïve Bayes classifier
Based on a joint probability model of the form:
𝐾
𝑝 𝑦, 𝒙 = 𝑝(𝑦)
𝑝(𝑥𝑘 |𝑦)
𝑘=1
Proof
𝐾
𝑝 𝑦, 𝒙 = 𝑝(𝑦)
𝑝(𝑥𝑘 |𝑦)
𝑘=1
Proof:
𝑝 𝑦, 𝒙 = 𝑝
=𝑝
=𝑝
=𝑝
=𝑝
=𝑝
𝑦, 𝑥1 , 𝑥2 , … 𝑥𝑘
𝑦 𝑝(𝑥1 , 𝑥2 , … 𝑥𝑘 |𝑦) all the features are independent
𝑦 𝑝(𝑥1 |𝑦)𝑝(𝑥2 , … 𝑥𝑘 |𝑦, 𝑥1 )
𝑦 𝑝 𝑥1 𝑦 𝑝 𝑥2 𝑦, 𝑥1 … 𝑝(𝑥𝑘 |𝑦, 𝑥1 , 𝑥2 , … , 𝑥𝑘 )
𝑦 𝑝 𝑥1 𝑦 𝑝 𝑥2 𝑦 … 𝑝 𝑥𝑘 𝑦
𝑦 𝐾
𝑘=1 𝑝 𝑥𝑘 𝑦
Naïve Bayes Classifer
Written as a factor graph
1
𝑝 𝒙, 𝒚 =
𝑍
𝐾
Ψ𝐴 (𝒙𝐴 , 𝒚A )
𝐴
𝑝 𝑦, 𝒙 = 𝑝(𝑦)
𝑝(𝑥𝑘 |𝑦)
𝑘=1
By defining Ψ 𝑦 = 𝑝(𝑦)
Ψ𝑘 𝑦, 𝑥𝑘 = 𝑝(𝑥𝑘 |𝑦)
However, the assumption is not practical, because the
features are not always independent
Ex. Classify the ball using following features:
color and weight V.S. size and weight
Logistic Regression
𝐾
log
𝑝
𝑦,
𝒙
=
log
𝑝
𝑦
+
𝑥𝑘 𝑦 of each
𝑘=1 log
Assumption that the log probability,
log𝑝𝑝(𝑦|𝐱),
𝐾
class
logis𝑝a 𝑦linear
𝐱 =function
−log Z of
𝑥 x,
+plus
𝜆𝑦 +a normalization
𝑗=1 𝜆𝑦,𝑗 𝑥𝑗
constant.
The conditional distribution:
1
𝑝 𝑦𝐱 =
exp 𝜆𝑦 +
𝑍(𝐱)
𝐾
𝜆𝑦,𝑗 𝑥𝑗
𝑗=1
𝑍 𝐱 = 𝑦 exp 𝜆𝑦 + 𝐾
𝑗=1 𝜆𝑦,𝑗 𝑥𝑗 is a normalizing constant
𝜆𝑦 : a bias weight that act as log 𝑝(𝑦) in naïve Bayes.
Different Notation
Using a set of feature functions
Defined for the feature weights: 𝑓𝑦′ ,𝑗 𝑦, 𝐱 = 𝟏 𝑦′ =𝑦 𝑥𝑗
Defined for the bias weights: 𝑓𝑦′ 𝑦, 𝐱 = 𝟏 𝑦′ =𝑦
𝑓𝑘 to index each feature function 𝑓𝑦′ ,𝑗
𝜆𝑘 to index corresponding weight 𝜆𝑦′ ,𝑗
1
𝑝 𝑦𝐱 =
exp
𝑍 𝐱
𝐾
𝜆𝑘 𝑓𝑘 (𝑦, 𝐱)
𝑘=1
Different Notation
𝐾
1
𝑝 𝑦𝐱 =
exp
𝑍 𝐱
𝜆𝑘 𝑓𝑘 (𝑦, 𝐱)
𝑘=1
1
𝑝 𝑦𝐱 =
exp 𝜆𝑦 +
𝑍(𝐱)
Where 𝜆1 𝑓1 𝑦, 𝐱 = 𝜆𝑦,1 𝑥1 +
𝜆2 𝑓2 𝑦, 𝐱 = 𝜆𝑦,2 𝑥2 +
𝜆𝑦
𝑘
𝜆𝑦
𝑘
.
.
.
𝜆𝐾 𝑓𝐾 𝑦, 𝐱 = 𝜆𝑦,𝐾 𝑥𝐾 +
𝜆𝑦
𝑘
𝐾
𝜆𝑦,𝑗 𝑥𝑗
𝑗=1
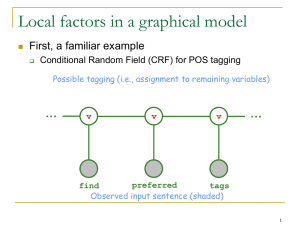
Sequence Models
Discussing the simplest form of dependency, in which the
output variables are arranged in a sequence
Use the hidden Markov model (HMM)
An HMM models a sequence of observations 𝑿 = 𝑥𝑡
𝑇
𝑡=1
by assuming there is an underlying sequence of states 𝒀 =
𝑦𝑡 𝑇𝑡=1 drawn from a finite state set S
Sequence Models
HMM makes two independence assumption:
First, each state 𝑦𝑡 is independent of all its ancestors 𝑦1 , 𝑦2 ,…, 𝑦𝑡−2
given its previous state 𝑦𝑡−1
Each observation variable 𝑥𝑡 depends only on the current state 𝑦𝑡
With these assumption, specify an HMM using three
probability distributions:
The distribution 𝑝(𝑦1 ) is over initial states
The transition distribution 𝑝(𝑦𝑡 |𝑦𝑡−1 )
The observation distribution 𝑝(𝑥𝑡 |𝑦𝑡 )
The form of HMM:
𝑇
𝑝 𝒚, 𝒙 =
𝑝 𝑦𝑡 𝑦𝑡−1 𝑝 𝑥𝑡 𝑦𝑡
𝑡=1
Initial state distribution 𝑝 𝑦1 = 𝑝 𝑦1 𝑦0
Proof
𝑇
𝑝 𝒚, 𝒙 =
𝑝 𝑦𝑡 𝑦𝑡−1 𝑝 𝑥𝑡 𝑦𝑡
𝑡=1
Proof :
𝑝 𝒚, 𝒙
each state 𝑦𝑡 is independent of all its ancestors
𝑦1 , 𝑦2 ,…, 𝑦𝑡−2 given its previous state 𝑦𝑡−1
Each observation variable 𝑥𝑡 depends only on
the current state 𝑦𝑡
=𝑝 𝒚 𝑝 𝐱𝐲
= 𝑝 𝑦1 , 𝑦2 , … , 𝑦𝑇 𝑝(𝑥1 , 𝑥2 , … , 𝑥𝑇 |𝒚)
= 𝑝 𝑦1 )𝑝(𝑦2 , … , 𝑦𝑇 |𝑦1
𝑝 𝑥1 𝑝(𝑥2 , … , 𝑥𝑇 |𝒚, 𝑥1 )
= 𝑝 𝑦1 )𝑝 𝑦2 𝑦1 … 𝑝(𝑦𝑇 |𝑦𝑡−1 , 𝑦𝑡−2 , … , 𝑦1
𝑝 𝑥1 𝑝 𝑥2 𝒚, 𝑥1 … 𝑝(𝑥𝑇 |𝒚, 𝑥𝑡−1 , 𝑥𝑡−2 , … 𝑥1 )
= 𝑝 𝑦1 )𝑝 𝑦2 𝑦1 … 𝑝(𝑦𝑇 |𝑦𝑡−1
𝑇
=
𝑝 𝑦𝑡 𝑦𝑡−1 𝑝 𝑥𝑡 𝑦𝑡
𝑡=1
𝑝 𝑥1 𝑝 𝑥2 𝑦2 … 𝑝(𝑥𝑇 |𝑦𝑇 )
Outline
2. Graphical Models
2-1 Definitions
2-2 Application of Graphical Models
2-3 Discriminative and Generative Models
Discriminative and Generative Models
Naïve Bayes is generative, meaning that it is based on a
model of the joint distribution 𝑝(𝑦, 𝒙)
Logistic regression is discriminative, meaning that it is
based on a model of the conditional distribution 𝑝(𝑦|𝒙)
The main difference is that the conditional distribution
does not include the distribution of 𝑝(𝒙)
To include interdependent features in a generative models,
two choice are used:
Enhancing the model to present dependencies among inputs
Making simplifying independence assumptions
Discriminative and Generative Models
If the two models are defined in the same hypothesis, the
two models can be converted with each other
Interpret it generatively as
exp 𝑘 𝜆𝑘 𝑓𝑘 𝑦, 𝐱
𝑝 𝑦, 𝐱 =
𝑘 𝜆𝑘 𝑓𝑘 𝑦, 𝐱
𝑦,𝐱 exp
Naïve Bayes and logistic regression form a generative-
discriminative pair
The principal advantage of discriminative modeling is that
it is better suited to including rich, overlapping features.
Discriminative and Generative Models
If the two models are defined in the same hypothesis, the
two models can be converted with each other
Interpret it generatively as
exp 𝑘 𝜆𝑘 𝑓𝑘 𝑦, 𝐱
𝑝 𝑦, 𝐱 =
𝑘 𝜆𝑘 𝑓𝑘 𝑦, 𝐱
𝑦,𝐱 exp
Naïve Bayes and logistic regression form a generative-
discriminative pair
The principal advantage of discriminative modeling is that
it is better suited to including rich, overlapping features.
Outline
3. Linear-Chain Conditional Random Fields
3-1 From HMMs to CRFs
3-2 Parameter Estimation
3-3 Inference
From HMMs to CRFs
Begin by considering the conditional distribution 𝑝(𝐲|𝐱) that
follows from the joint distribution 𝑝 𝒚, 𝒙 of an HMM
Key point: with a particular choice of feature functions
Sequence HMM joint distribution:
𝑇
𝑝 𝒚, 𝒙 =
𝑝 𝑦𝑡 𝑦𝑡−1 𝑝 𝑥𝑡 𝑦𝑡
𝑡=1
Rewriting it generally:
𝑝 𝒚, 𝒙
1
= exp
𝑍
𝜆𝑖𝑗 𝟏 𝑦𝑡 =𝑖 𝟏 𝑦𝑡−1=𝑗 +
𝑡 𝑖,𝑗∈𝑆
𝜇𝑜𝑖 1 𝑦𝑡 =𝑖 1 𝑥𝑡=𝑜
𝑡
𝑖∈𝑆 𝑜∈𝑂
Reviewing The HMM
HMM makes two independence assumption:
First, each state 𝑦𝑡 is independent of all its ancestors 𝑦1 , 𝑦2 ,…, 𝑦𝑡−2
given its previous state 𝑦𝑡−1
Each observation variable 𝑥𝑡 depends only on the current state 𝑦𝑡
With these assumption, specify an HMM using three
probability distributions:
The distribution 𝑝(𝑦1 ) is over initial states
The transition distribution 𝑝(𝑦𝑡 |𝑦𝑡−1 )
The observation distribution 𝑝(𝑥𝑡 |𝑦𝑡 )
The form of HMM:
𝑇
𝑝 𝒚, 𝒙 =
𝑝 𝑦𝑡 𝑦𝑡−1 𝑝 𝑥𝑡 𝑦𝑡
𝑡=1
Initial state distribution 𝑝 𝑦1 = 𝑝 𝑦1 𝑦0
From HMMs to CRFs
HMM joint distribution: 𝑝 𝒚, 𝒙 =
𝑇
𝑡=1 𝑝
𝑦𝑡 𝑦𝑡−1 𝑝 𝑥𝑡 𝑦𝑡
Rewriting it generally :
𝑝 𝒚, 𝒙
1
= exp
𝑍
𝜆𝑖𝑗 𝟏 𝑦𝑡 =𝑖 𝟏 𝑦𝑡−1=𝑗 +
𝑡 𝑖,𝑗∈𝑆
S: the set of transition states
𝜆𝑖𝑗 : log 𝑝(𝑦𝑡 = 𝑖|𝑦𝑡−1 = 𝑗)
O: the set of observation states
𝜇𝑜𝑖 : log 𝑝(𝑥𝑡 = 𝑖|𝑦𝑡 = o)
Easy to use the feature functions
𝜇𝑜𝑖 1 𝑦𝑡 =𝑖 1 𝑥𝑡=𝑜
𝑡
𝑖∈𝑆 𝑜∈𝑂
HMM by Using Feature Functions
Each feature function has the form 𝑓𝑘 𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡
𝑓𝑖𝑗 𝑦, 𝑦 ′ , 𝑥 = 𝟏 𝑦=𝑖 𝟏{𝑦′ =𝑗} for each transition (𝑖, 𝑗)
𝑓𝑖𝑜 𝑦, 𝑦 ′ , 𝑥 = 𝟏 𝑦=𝑖 𝟏{𝑥=𝑜} for each state-observation
pair (𝑖, 𝑗)
Write an HMM as :
1
𝑝 𝒚, 𝒙 = exp
𝑍
𝐾
𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝑘=1
Compare
1
𝑝 𝒚, 𝒙 = exp
𝑍
1
𝑝 𝒚, 𝒙 = exp
𝑍
𝐾
𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝑘=1
𝜆𝑖𝑗 𝟏 𝑦𝑡 =𝑖 𝟏 𝑦𝑡−1 =𝑗 +
𝑡 𝑖,𝑗∈𝑆
𝜇𝑜𝑖 1 𝑦𝑡 =𝑖 1 𝑥𝑡 =𝑜
𝑡
𝑖∈𝑆 𝑜∈𝑂
Let
𝜆𝑘 =
𝜇𝑜𝑖 ,
𝜆𝑖𝑗 ,
𝑘 = 1~𝑇|𝑆|2
𝑘 = 𝑇 𝑆 2 + 1~𝑇 𝑆 2 + 𝑇|𝑆||𝑂|
𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
=
𝟏 𝑦𝑡=𝑖 𝟏 𝑦𝑡−1=𝑗 ,
𝑘 = 1~𝑇|𝑆|2
1 𝑦𝑡 =𝑖 1 𝑥𝑡 =𝑜 , 𝑘 = 𝑇 𝑆 2 + 1~𝑇 𝑆 2 + 𝑇|𝑆||𝑂|
Linear-Chain CRF
By the definition of the conditional distribution:
𝑝(𝒚, 𝒙)
𝑝 𝒚𝒙 =
𝑝(𝒚, 𝒙)𝑝(𝒚)
𝑝 𝒚|𝒙 =
Use the discussion of the joint
distribution:
𝑝(𝒚)
𝐾
𝑝(𝒚,
𝒙)
1
𝑝=
𝒚, 𝒙 = exp ′
𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝑍𝑝(𝑦 , 𝒙)
′
𝑥
𝑘=1
exp 𝐾
𝑘=1 𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
And obtain the
=distribution 𝑝 𝐾𝒚 by 𝑝 𝒚, 𝒙
𝐾 𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝑥 ′ exp 𝑘=1
𝑝 𝒚 =
𝑥′
1
exp
𝑍
𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝑘=1
Linear-Chain CRF
Let the parameter 𝑥𝑡 of the feature function
𝑓𝑘 𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 be 𝒙𝒕
Lead to the general definition of linear-chain CRF
Definition 1.1
𝐘, 𝐗: random vectors
Λ = {𝜆𝑘 } ∈ 𝑅 𝐾 : a parameter vector
{𝑓𝑘 (𝑦, 𝑦 ′ , 𝑥𝑡 }𝐾
𝑘=1 : a set of real-valued feature functions
linear-chain conditional random field
1
𝑝 𝒚𝒙 =
exp
𝑍(𝒙)
𝑍 𝒙 =
𝑦 exp
𝐾
𝜆𝑘 𝑓𝑘 𝑦𝑡 , 𝑦𝑡−1 , 𝒙𝒕
𝑘=1
𝐾
𝑘=1 𝜆𝑘 𝑓𝑘
𝑦𝑡 , 𝑦𝑡−1 , 𝒙𝒕
Linear-Chain CRF
HMM-liked CRF:
exp
𝑝 𝒚𝒙 =
𝑦 ′ exp
𝐾
𝑘=1 𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝐾
𝑘=1 𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
linear-chain CRF:
1
𝑝 𝒚𝒙 =
exp
𝑍(𝒙)
𝐾
𝜆𝑘 𝑓𝑘 𝑦𝑡 , 𝑦𝑡−1 , 𝒙𝒕
𝑘=1
Linear-Chain CRF
Allow the score of the transition (𝑖, 𝑗) to depend on the
current observation vector, by adding a feature
1{𝑦𝑡 =𝑗} 1{𝑦𝑡−1 =1} 1{𝑥𝑡 =0}
𝑝 𝒚𝒙 =
1
exp
𝑍(𝒙)
𝐾
𝑘=1 𝜆𝑘 𝑓𝑘
𝑦𝑡 , 𝑦𝑡−1 , 𝒙𝒕
However, the normalization constant sums over all
possible state sequence, an exponentially large number of
terms.
It can be computed efficiently by forward-backward, as
explaining it later
Outline
3. Linear-Chain Conditional Random Fields
3-1 From HMMs to CRFs
3-2 Parameter Estimation
3-3 Inference
Parameter Estimation
Discuss how to estimate the parameter Λ = 𝜆𝑘
Given iid training data 𝒟 = {𝒙 𝑖 , 𝒚(𝑖) }𝑁
𝑖=1 ,
where 𝒙
𝑖
𝑖
𝑖
𝑖
= {𝑥1 , 𝑥2 , … , 𝑥 𝑇 } is a sequence of input
𝑖
𝑖
𝑖
and 𝒚 𝑖 = {𝑦1 , 𝑦2 , … , 𝑦𝑇 } is a sequence of the desired
prediction
Performed by penalized maximum likelihood
Because modeling the conditional distribution, called the
conditional log likelihood, is appropriate:
𝑁
𝑙𝑜𝑔 𝑝(𝒚(𝑖) |𝒙(𝑖) )
ℓ 𝜃 =
𝑖=1
Parameter Estimation
After substituting in the CRF model into the likelihood
𝑁
𝑇
𝐾
𝑁
𝑖
ℓ 𝜃 =
𝑖
𝑖
log 𝑍(𝐱 𝑖 )
𝜆𝑘 𝑓𝑘 (𝑦𝑡 , 𝑦𝑡−1 , 𝒙𝑡 ) −
𝑖=1 𝑡=1 𝑘=1
𝑖=1
As a measure to avoid overfitting, use regularization, which is a
penalty on weight vectors whose norm is too large
A common choice of penalty is based on the Euclidean norm of
𝜃 and on a regularization parameter 1/2σ2
Regularized log likelihood:
𝑁
𝑇
𝐾
ℓ 𝜃 =
𝑁
𝜆𝑘 𝑓𝑘 (𝑦𝑡
𝑖=1 𝑡=1 𝑘=1
𝑖
𝑖
𝑖
, 𝑦𝑡−1 , 𝒙𝑡
𝐾
log 𝑍(𝐱 𝑖 ) −
)−
𝑖=1
𝑘=1
𝜆2𝑘
2σ2
Parameter Estimation
The function ℓ 𝜃 cannot be maximized in closed form.
The partial differential:
𝜕ℓ
=
𝜕𝜆𝑘
𝑁
𝑇
𝑓𝑘 (𝑦𝑡
𝑖
𝑖
𝑖
, 𝑦𝑡−1 , 𝒙𝑡
𝑁
𝑇
𝐾
𝑖
𝑓𝑘 𝑦, 𝑦 ′ , 𝒙𝑡
)−
𝑖
𝑝(𝑦, 𝑦 ′ |𝒙
𝑖=1 𝑡=1 𝑦,𝑦 ′
𝑖=1 𝑡=1
)−
𝑘=1
First term: the expect value of 𝑓𝑘 under the empirical
distribution 𝑝 𝒚, 𝒙 =
1
𝑁
𝑁
𝑖=1 1{𝒚=𝒚(𝑖) } 1{𝒙=𝒙(𝑖) }
Second term: arises from the derivative of log Z 𝐱 , the
expectation of 𝑓𝑘 under the model distribution 𝑝(𝒚|𝒙; 𝜃)𝑝(𝒙)
𝜆𝑘
σ2
Optimize ℓ 𝜃
The function ℓ 𝜃 is concave, which follows from the convexity
of functions of the form 𝑔 𝐱 = 𝑙𝑜𝑔
𝑖 exp 𝑥𝑖
Every local optimum is also a global optimum in concave
functions
Adding regularization ensures ℓ is strictly concave, which
implies that it has exactly one global optimum
The simplest approach to optimize ℓ is steepest ascent along the
gradient.
Newton’s method converges much faster because it takes into
the curvature of the likelihood, but it requires computing the
Hessian
Optimize ℓ 𝜃
Quasi-Newton methods: BFGS [Bertsekas,1999]
Limited-memory version of BFGS, due to Byrd et al. [1994]
When such second-order methods are used, gradient-based
optimization is much faster than original approaches based on
iterative scaling in Lafferty et al. [2001] as shown
experimentally by several authors [Sha and Pereira, 2003,
Wallcach, 2002, Malouf, 2002, Minka, 2003]
Computational cost:
𝑝(𝑦𝑡 , 𝑦𝑡−1 |𝐱): 𝑂(𝑇𝑀2 ) where M is the number of state of each 𝑦𝑡
Total computer cost: 𝑂(𝑇𝑀2 𝑁𝐺) where N is the number of train
examples and G is the number of gradient computions
Outline
3. Linear-Chain Conditional Random Fields
3-1 From HMMs to CRFs
3-2 Parameter Estimation
3-3 Inference
Inference
Two common inference problems for CRFS:
During training, computing the gradient requires marginal
distribution for each edge 𝑝 𝑦𝑡 , 𝑦𝑡−1 𝒙 , and computing the
likelihood requires 𝑍(𝒙)
To label an unseen instance, we compute the most likely labeling
𝒚∗ = arg max 𝑝(𝐲|𝐱)
𝒚
In linear-chain CRFs, inference tasks can be performed
efficiently and exactly by dynamic-programming algorithms for
HMMs
Here, review the HMM algorithms, and extend them to linear
CRFs
Introduce Notations
HMM: 𝑝 𝒚, 𝒙 =
𝑇
𝑡=1 𝑝
𝑦𝑡 𝑦𝑡−1 𝑝 𝑥𝑡 𝑦𝑡
Viewed as a factor graph: 𝑝 𝒙, 𝒚 =
1
𝑍
𝑡 Ψ𝑡 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
Define the factors and normalization constant as :
𝑍=1
Ψ𝑡 𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 ≝ 𝑝 𝑦𝑡 = 𝑗 𝑦𝑡−1 = 𝑖 𝑝 𝑥𝑡 = 𝑥 𝑦𝑡 = 𝑗
If viewed as a weighted finite state machine, the Ψ𝑡 𝑗, 𝑖, 𝑥 is the
weight on the transition from state 𝑖 to state 𝑗, when the current
observation is 𝑥
HMM Forward Algorithm
Used to compute the probability 𝑝(𝐱) of the observations
First, Using the distributive law:
𝑝 𝒙
=
𝑝(𝒙, 𝒚)
𝒚
=
Ψ𝑡 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝒚
𝑡
=
Ψ𝑡 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝑦1 ,𝑦2 ,…,𝑦𝑇
=
𝑡
Ψ𝑇 (𝑦𝑇 , 𝑦𝑇−1 , 𝑥 𝑇 )
𝑦𝑇 𝑦𝑇−1
Ψ𝑇−1 (𝑦𝑇−1 , 𝑦𝑇−2 , 𝑥 𝑇−1 ) …
𝑦𝑇−2
HMM Forward Algorithm
𝑝 𝒙 =
Ψ𝑇 (𝑦𝑇 , 𝑦𝑇−1 , 𝑥 𝑇 )
𝑦𝑇 𝑦𝑇−1
Ψ𝑇−1 (𝑦𝑇−1 , 𝑦𝑇−2 , 𝑥 𝑇−1 ) …
𝑦𝑇−2
Each of the intermediate sums is reused many times, and we can
save an exponential amount of work by caching the inner sums
forward variables 𝛼𝑡 :
Each is a vector of size M (the number of states)
Store the intermediate sums
HMM Forward Algorithm
𝑝 𝒙 =
Ψ𝑇 (𝑦𝑇 , 𝑦𝑇−1 , 𝑥 𝑇 )
𝑦𝑇 𝑦𝑇−1
Ψ𝑇−1 (𝑦𝑇−1 , 𝑦𝑇−2 , 𝑥 𝑇−1 ) …
𝑦𝑇−2
Defined as :
𝛼𝑡 𝑗 ≝ 𝑝 𝒙 1…𝑡 , 𝑦𝑡 = 𝑗
𝑡−1
=
Ψ𝑡 (𝑗, 𝑦𝑡−1 , 𝑥𝑡 )
𝒚 1…𝑡
Ψ𝑡 ′ (𝑦𝑦′ , 𝑦𝑦′−1 , 𝑥𝑡′ )
𝑡 ′ =1
Compute by the recursion:
𝛼𝑡 𝑗 =
𝑖∈𝑠 Ψ𝑡 (𝑗, 𝑖, 𝑥𝑡 ) 𝛼𝑡−1
Initialization: 𝛼1 𝑗 =
𝑖
𝑦𝑇 𝛼 𝑇
𝑦𝑇
HMM Forward Algorithm
𝑝 𝒙 =
Ψ𝑇 (𝑦𝑇 , 𝑦𝑇−1 , 𝑥 𝑇 )
𝑦𝑇 𝑦𝑇−1
Ψ𝑇−1 (𝑦𝑇−1 , 𝑦𝑇−2 , 𝑥 𝑇−1 ) …
𝑦𝑇−2
Backward :
𝛽𝑡 𝑗 ≝ 𝑝 𝒙 𝑡+1…𝑇 |𝑦𝑡 = 𝑖
𝑇
=
Ψ𝑡 ′ (𝑦𝑦′ , 𝑦𝑦′−1 , 𝑥𝑡′ )
𝑦 𝑡+1…𝑇 𝑡 ′ =𝑡+1
Recursion:
𝛽𝑡 𝑖 =
𝑗∈𝑆 Ψ𝑡+1 (𝑗, 𝑖, 𝑥𝑡+1 )𝛽𝑡+1 (𝑗)
Initialization: 𝛽𝑇 𝑖 = 1
HMM Forward Algorithm
Appling the distributive law:
𝑝 𝑦𝑡−1 , 𝑦𝑡 𝐱
𝑡−1
= Ψ𝑡 𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡
Ψ𝑡 ′ 𝑦𝑡 ′ , 𝑦𝑡 ′ −1 , 𝑥𝑡 ′
𝑦 1…𝑡−2 𝑡 ′ =1
𝑇
Ψ𝑡 ′ (𝑦𝑡 ′ , 𝑦𝑡 ′ −1 , 𝑥𝑡 ′ )
𝑦 𝑡+1…𝑇 𝑡 ′ =𝑡+1
Recursion:
𝑝 𝑦𝑡−1 , 𝑦𝑡 𝐱 ∝ 𝛼𝑡−1 𝑦𝑡−1 Ψ𝑡 𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 𝛽𝑡 𝑦𝑡
HMM Forward Algorithm
Finally, compute the globally most probable assigment:
y ∗ = arg max 𝑝(𝑦|𝑥)
𝒚
This yields the Viterbi recursion if all the summations are
replaced by maximization:
𝛿𝑡 𝑗 = max Ψ𝑡 (𝑗, 𝑖, 𝑥𝑡 ) 𝛿𝑡−1 𝑖
𝑖∈𝑠
Forward-Backward Algorithm For CRF
The definition forward recursion, the backward recursion, and
the Viterbi recursion of CRF are the same as HMM
Use the recursion to computer 𝑍 𝐱
𝐾
𝑍 𝒙 =
exp
𝐾
𝑦
=
𝜆𝑘 𝑓𝑘 𝑦𝑡 , 𝑦𝑡−1 , 𝒙𝒕
𝑘=1
exp 𝜆𝑘 𝑓𝑘 𝑦𝑡 , 𝑦𝑡−1 , 𝒙𝒕
𝑦 𝑘=1
𝑝 𝒙
=
Ψ𝑡 (𝑦𝑡 , 𝑦𝑡−1 , 𝑥𝑡 )
𝒚
𝑡