Bruker DPX-300 NMR–Basic Operation Department of Chemistry, Tufts University
advertisement

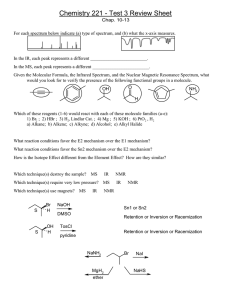
Department of Chemistry, Tufts University Bruker DPX-300 NMR–Basic Operation NOTE: These abbreviated operation notes are meant as a reminder to those who have been trained on the NMR. Contact the Instrument Specialist, David Wilbur (X72163) for training. Login 1. Sign the log book. 2. Enter username and password to log in. 3. Type xwinnmr in any command window to start the NMR application. Lock and Shim 1. Select Windows/Lock and Windows/BSMS Panel from the menu. Select SHIM from BSMS panel 2. In Shim panel LOCK OFF, SPIN OFF, then LIFT ON. Change sample (use depth gauge). LIFT OFF, SPIN ON 3. Type lock in command line, select solvent. 4. Adjust Z1, Z2 for maximum signal. Left click decreases value, middle click increases. Adjust LOCK GAIN to keep signal level 60-80% of maximum. Setup 1. Select File/New. Enter NAME and EXPNO. Select SAVE. 2. Select File/Copy/Parameter file from. Select PROTON or C13CPD for proton or 13C spectra. Select Copy All 3. Type eda. Select solvent, set PROSOL to true. Change value of NS if desired. Select SAVE Acquire Data 1. Type rga. 2. Type zg Process data 1. Type efp to transform and phase to last phase values 2. Select phase. Select biggest. Select and hold PH0, move mouse vertically for best phase at cursor. Select and hold PH1, move mouse vertically for best phase away from cursor. Integrate 1. Select Analysis/Auto integration. For narrow, well separated peaks, no more is needed. 2. Select Analysis/Manual integration. Select File/read intreg if previous integrals exist. 3. To define a new integral region, left click in the spectrum window, then middle click beginning and end of desired region. 4. To select an existing integral, left click in the spectrum window, then left click again anywhere in the desired integral region. A small arrow will mark the top of the integral. Use the current: buttons to calibrate or delete the integral. 5. Select return, then save as intrng and return to exit integration. Peak Picking 1. Type setmi to set the threshold. Type pps to list peaks on screen, pp to list on printer. Revised 2/28/06 D. Wilbur Department of Chemistry, Tufts University Plotting within xwinnmr 1. Select the Plot button. Ignore any warnings about missing files (select OK). Type view to preview the plot. Plotting with Xwinplot 1. Type xwinplot in the command line. 2. In the xwinplot menu select XWINNMR/XWINNMR Interface. Select Edit Title, Create Peak list and Create Parameter list. Select Close. 3. To create a spectrum, select the spectrum icon, then click and drag to the desired size. Multiple or overlapping spectra can be created on the same page. 4. To create title or parameter list, select the icon, and left click to place it. 5. To move an object, middle click and drag. 6. To edit object properties, select the Mark icon (little green squares), then select the object, then select the Edit button 7. To print, select File/Print. Exporting Data 1. Any output from xwinplot can be saved as a graphics file. In xwinplot select Options/Printer Setup from the menu. Select one of the graphics file formats at the end of the printer list. Available formats are EPSI (Encapsulated Postscript), PCX (Windows Paint), or TIFF. To print, select File/Print, then select Print to File. 2. Any spectrum can be saved as a text file containing point number, Hz, ppm and intensity values by typing convbin2asc. The result will written to the same directory as the original spectrum, ie /datanmr/data/<user name>/nmr/NAME/EXPNO/pdata/PROCNO/asciispec.txt 3. The NMR computer is accessible via FTP as iris.chem.tufts.edu. You will need to use your login and password to connect via FTP. When Finished 4. Remove your sample, replace it with one of the sealed samples. Lock and shim the sealed sample. 5. Select File/Exit from the menu to exit Xwinnmr. 6. Select Desktop/Log Out from the Toolchest menu. 7. Tidy up. Revised 2/28/06 D. Wilbur