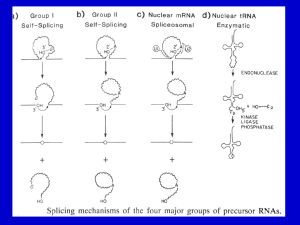
Splicing RNA: Mechanisms
advertisement

Splicing RNA: Mechanisms Splicing of Group I and II introns • Introns in fungal mitochondria, plastids, Tetrahymena pre-rRNA • Group I – Self-splicing – Initiate splicing with a G nucleotide – Uses a phosphoester transfer mechanism – Does not require ATP hydrolysis. • Group II – self-splicing – Initiate splicing with an internal A – Uses a phosphoester transfer mechanism – Does not require ATP hydrolysis Self-splicing in pre-rRNA in Tetrahymena : T. Cech et al. 1981 + Exon 1 Intron 1 Exon 2 Exon 1 Exon 2 Intron 1 •Products of splicing were resolved by gel electrophoresis: + + + pre-rRNA + Nuclear extract Additional proteins + + GTP + + are NOT needed for pre-rRNA Spliced exon Intron circle Intron linear splicing of this prerRNA! Do need a G nucleotide (GMP, GDP, GTP or Guanosine). Self-splicing by a phosphoester transfer mechanism G OH U P P Exon 1 U U Intron 1 P P A Exon 1 G U P P + Exon 2 G A P Exon 2 Intron 1 N15 N16 P G P OH G A N15 OH P + Circular intron A catalytic activity in Group I intron • Self-splicing uses the intron in a stoichiometric fashion. • But the excised intron can catalyze cleavage and addition of C’s to CCCCC Group I intron catalyzes cleavage and nucleotide addition 2 pCCCCC-OH pCCCC-OH + pCCCCCC-OH 3'G OH 5'pCCCCC-OH GGGAGG 3'G C-OH 5' G 3'C-OH 5'pCCCC-OH GGGAGG 5'pCCCCC-OH GGGAGG 5' 3'G OH 5' GGGAGG 5' + 5'pCCCCCC-OH The intron folds into a particular 3-D structure • Has active site for phosphoester transfer • Has G-nucleotide binding site Active sites in Group I intron self-splicing ex2 3' 3'G414 OH 414 G G-binding site G-OH 5' G ex1 5' Substrate binding site 3' CUCUCU GGGAGG 2nd transfer IGS UUUACCU GGGAGG ex1 + ex2 1st transfer ex2 3rd transfer G414 ex1 5' G 414 CUCUCU OH GGGAGG G 5' GGGAGG + 5' G UUUACCU Domains of the Group I intron ribozyme http://www.tulane.edu/~biochem/nolan/lectures/rna/grz.htm RNAs that function as enzymes • • • • • • RNase P Group I introns Group II introns rRNA: peptide bond formation Hammerhead ribozymes: cleavage snRNAs involved in splicing Hammerhead ribozymes • A 58 nt structure is used in self-cleavage • The sequence CUGA adjacent to stemloops is sufficient for cleavage 5' 3' AA A GGCC CCGG A CG U A C G AUC U G GU A Bond that is cleaved. ACCAC C UGGUG CUGA is required for catalysis Design hammerhead ribozymes to cleave target RNAs AA A substrate strand 5' GGCC enzyme strand 3' CCGG A UA G A CG U A C G AUC GU A GU Bond that is cleaved. ACCAC C UGGUG Potential therapy for genetic disease. Mechanism of hammerhead ribozyme • The folded RNA forms an active site for binding a metal hydroxide • Abstracts a proton from the 2’ OH of the nucleotide at the cleavage site. • This is now a nucleophile for attack on the 3’ phosphate and cleavage of the phosphodiester bond. Phosphotransfers for Group I vs. Group II & pre-mRNA Exon 1 3’ G HO 2’ Exon 1 Exon 2 HO Exon 2 2’ A G OH OH Exon 1+2 A Exon 1+2 + + G 2’ A OH Group I Group II and pre-mRNA Splicing of pre-mRNA • The introns begin and end with almost invariant sequences: 5’ GU…AG 3’ • Use ATP to assemble a large spliceosome • Mechanism is similar to that of the Group II fungal introns: – Initiate splicing with an internal A – Uses a phosphoester transfer mechanism for splicing Initiation of phosphoester transfers in pre-mRNA • Uses 2’ OH of an A internal to the intron • Forms a branch point by attacking the 5’ phosphate on the first nucleotide of the intron • Forms a lariat structure in the intron • Exons are joined and intron is excised as a lariat • A debranching enzyme cleaves the lariat at the branch to generate a linear intron • Linear intron is degraded Splicing of pre-mRNA, step 1 Splicing of pre-mRNA, step 2 Investigation of splicing intermediates In vitro splicing reaction: nuclear extracts + ATP+ labeled pre-mRNA Resolve reaction intermediates and products on gels. Some intermediates move slower than pre-mRNA. Suggest they are not linear. Use RNase H to investigate structure of intermediate. RNase H cuts RNA in duplex with RNA or DNA. RNase H RNA 5' 3' ||||| 5' oligodeoxyribonucleotide + RNase H + oligonucleotides complementary to different regions give very different products precursor RNA exon 1 5' intron exon 2 3' splicing reaction exons joined in a linear molecule+ excised intron, non-linear molecule 1 5' 2 3' 5' 3 Map of positions of oligodeoxyribonucleotides that annealed to different regions of the excised intron. This is not the structure of the excised intron. 4 RNase H 3' 3' 5' 3' + 3' 5' Ans wer: intron exon 2 3' s plicing reaction exons joined in a linear m olecule + excis ed intron, non-linear m olecule 1 2 3 Map of pos itions of oligodeoxyribonucleotides that annealed to different regions of the excis ed intron. 4 2 GU Analysis reveals a lariate structure in intermediate precurs or RNA exon 1 5' A 3 1 AG 4 oligo 1 oligo 2 oligo 4 3' 5' oligo 3 3' After annealing with the oligo, the heteroduplexes were treated with RNas e H 3' 5' 3' + 3' 5' Involvement of snRNAs and snRNPs • snRNAs = small nuclear RNAs • snRNPs = small nuclear ribonucleoprotein particles • Antibodies from patients with the autoimmune disease systemic lupus erythematosus (SLE) can react with proteins in snRNPs – Sm proteins • Addition of these antibodies to an in vitro premRNA splicing reaction blocked splicing. • Thus the snRNPs were implicated in splicing snRNPs • U1, U2, U4/U6, and U5 snRNPs – Have snRNA in each: U1, U2, U4/U6, U5 – Conserved from yeast to human – Assemble into spliceosome – Catalyze splicing • Sm proteins bind “Sm RNA motif” in snRNAs – 7 Sm proteins: B/B’, D1, D2, D3, E, F, G – Each has similar 3-D structure: alpha helix followed by 5 beta strands – Sm proteins interact via beta strands, may form circle around RNA Sm proteins may form ring around snRNAs ANGUS I. LAMOND Nature 397, 655 - 656 (1999) RNA splicing: Running rings around RNA Predicted structure of assembled Sm proteins 4th beta strand of one Sm protein interacts with 5th beta strand of next. Channel for single strand of RNA ANGUS I. LAMOND Nature 397, 655 - 656 (1999) RNA splicing: Running rings around RNA Assembly of spliceosome • The spliceosome is a large protein-RNA complex in which splicing of pre-mRNAs occurs. • snRNPs are assembled progressively into the spliceosome. – – – – U1 snRNP binds (and base pairs) to the 5’ splice site U2 snRNP binds (and base pairs) to the branch point U4-U6 snRNP binds, and U4 snRNP dissociates U5 snRNP binds • Assembly requires ATP hydrolysis • Assembly is aided by various auxiliary factors and splicing factors. Spliceosome assembly and catalysis U2 snRNP 2’ A HO Exon 1 U1 snRNP 2’ A 2’ HO Exon 2 A HO Sm proteins U6 U5 snRNP Other proteins U4/U6 snRNP snRNAs U G O H U4 A U6 U2 Exons 1+2 U1 U 2’ A G O H U6 U4? 2’ A U5 Spliceosome Catalysis by U6/U2 on branch oligonucleotide in vitro Figure 1 Base-pairing interactions in the in vitro-assembled complex of U2–U6 and the branch oligonucleotide (Br). Shaded boxes mark the invariant regions in U6 and previously established base-paired regions are indicated. Dashed lines connect psoralen-crosslinkable nucleotides (S.V. and J.L.M., unpublished data). The circled residues connected by a zigzag can be crosslinked by ultraviolet light. The underlined residues in Br constitute the yeast branch consensus sequence. Asterisks denote the residues involved in the covalent link between Br and U6 in RNA X (see text). Arrowheads point to residues involved in a genetically proven interaction in yeast22. Numbers indicate nucleotide positions from the 5' ends of full-length human U2 and U6. Nature 413, 701 - 707 (2001) Splicing-related catalysis by protein-free snRNAs SABA VALADKHAN & JAMES L. MANLEY RNA editing • RNA editing is the process of changing the sequence of RNA after transcription. • In some RNAs, as much as 55% of the nucleotide sequence is not encoded in the (primary) gene, but is added after transcription. • Examples: mitochondrial genes in trypanosomes and Leishmania. • Can add, delete or change nucleotides by editing Addition of nucleotides by editing • Uses a guide RNA that is encoded elsewhere in the genome • Part of the guide RNA is complementary to the mRNA in vicinity of editing • U nt at the the 3’ end of the guide RNA initiates a series of phosphoester transfers that result in insertion of that U at the correct place. • More U’s are added sequentially at positions directed by the guide RNA • Similar mechanism to that used in splicing What is a gene? • Making a correctly edited mRNA requires one segment of DNA to encode the initial transcript and a different segment of DNA to encode each guide RNA. • Thus making one mRNA that uses 2 guide RNAs requires 3 segments of DNA - is this 3 genes or 1 gene? • Loss-of-function mutations in any of those 3 DNA segments result in an nonfunctional product (enzyme), but they will complement in trans in a diploid analysis! • This is an exception to the powerful cis-trans complementation analysis to define genes. Mammalian example of editing • Apolipoprotein B in the intestine is much shorter than apolipoprotein B in the liver. • They are encoded by the same gene. • The difference results from a single nt change in codon 2153: • CAA for Gln in liver, but UAA for termination of translation in intestine • The C is converted to U in intestine by a specific deaminating enzyme, not by a guide RNA.