1. The statistical analysis result for verifying the statistical significance between clusters.
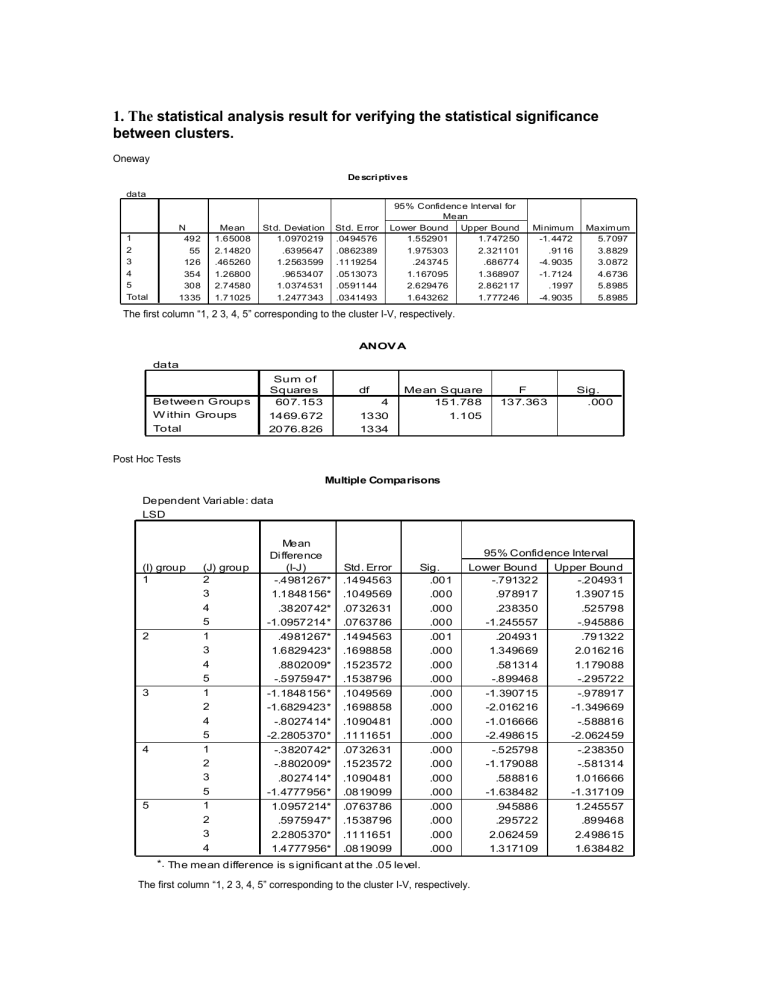
1. The statistical analysis result for verifying the statistical significance between clusters.
Oneway
De scri ptives data
1
2
3
4
5
Total
N
492
55
126
354
308
1335
Mean
1.65008
2.14820
.465260
1.26800
2.74580
1.71025
95% Confidenc e Int erval for
Mean
St d. Deviat ion St d. E rror Lower Bound Upper Bound
1.0970219
.0494576
1.552901
1.747250
.6395647
.0862389
1.2563599
.1119254
.9653407
.0513073
1.975303
.243745
1.167095
2.321101
.686774
1.368907
1.0374531
.0591144
1.2477343
.0341493
2.629476
1.643262
2.862117
1.777246
The first column “1, 2 3, 4, 5” corresponding to the cluster I-V, respectively.
Mi nimum Maxim um
-1. 4472 5.7097
.9116
-4. 9035
-1. 7124
3.8829
3.0872
4.6736
.1997
-4. 9035
5.8985
5.8985
ANOV A data
Between Groups
W ithin Groups
Total
Post Hoc Tests
Sum of
Squares
607.153
1469.672
2076.826
df
4
1330
1334
Mean S quare
151.788
1.105
F
137.363
Sig.
.000
Multiple Comparisons
Dependent Vari able: data
LSD
(I) group
1
2
3
4
5
(J) group
2
3
4
5
1
3
4
5
1
2
4
5
1
2
3
5
1
2
3
4
Mean
Di fference
(I-J) Std. Error
-.4981267* .1494563
1.1848156* .1049569
.3820742* .0732631
-1.0957214* .0763786
.4981267* .1494563
1.6829423* .1698858
.8802009* .1523572
-.5975947* .1538796
-1.1848156* .1049569
-1.6829423* .1698858
-.8027414* .1090481
-2.2805370* .1111651
-.3820742* .0732631
-.8802009* .1523572
.8027414* .1090481
-1.4777956* .0819099
1.0957214* .0763786
.5975947* .1538796
2.2805370* .1111651
1.4777956* .0819099
*. The mean difference is s igni ficant at the .05 level.
Sig.
.001
.000
.000
.000
.001
.000
.000
.000
.000
.000
.000
.000
.000
.000
.000
.000
.000
.000
.000
.000
The first column “1, 2 3, 4, 5” corresponding to the cluster I-V, respectively.
95% Confidence Interval
Lower Bound
-.791322
Upper Bound
-.204931
.978917
.238350
-1.245557
.204931
1.390715
.525798
-.945886
.791322
1.349669
.581314
-.899468
-1.390715
-2.016216
-1.016666
-2.498615
-.525798
-1.179088
.588816
-1.638482
.945886
.295722
2.062459
1.317109
2.016216
1.179088
-.295722
-.978917
-1.349669
-.588816
-2.062459
-.238350
-.581314
1.016666
-1.317109
1.245557
.899468
2.498615
1.638482
2 Validation of microarray data by real-time RT-PCR.
In order to verify the microarray result, the relative expression levels of 8 genes at several time points (0, 4, 10, and 15) were estimated by Quantitative real-time RT-PCR.
Gene-specific primers were designed for the genes of interest and the 18S rRNA using
Primer Express software (Applied Biosystems) and are shown in Table 1.
Table 1 Gene-specific primers used for real-time RT-PCR assays
Target Primer Sequence a
F,5'- CGCTGGCTTCTTAGAGGGACTAT -3'
18S rRNA
R,5'- TGCCTCAAACTTCCATCGACTT -3'
DW679821
DW694001
DW698978
DW691154
DW699406
F,5’- GAGGTGTTTATCTTTTCGCTGTC --3’
R,5’- AGGTTTGTATTTGGGGTATCC --3’
F,5’- ATCAAGGAACAGAAGCAACG --3’
R,5’- TGGAAGGTGGGCAGAGTAA --3’
F,5’- CCCATCCCGAGTTATTTCC --3’
R,5’- TTTACCCATACGCTTCATCAG --3’
F,5’- AACCTGACGAGCAAACCAA --3’
R,5’- AATGACAACAGAGGCGATAAAG --3’
F,5’- CCTTTTAGGTTCCCGCTGAG --3’
R,5’- GGCAAATAACAACAACGCAAG --3’
DW699957
DW699255
DW700354
DW702524
DW683219
F, 5’- AAGTTTCCGCCAATGCCA --3’
R,5’- ACCCTTCAATGCGTCCAG --3’
F, 5’- CCCACCAGAATAACAGATGC --3’
R,5’- CAGTGATGTGAACTCCGAGCT --3’
F, 5’- GAGACGCCGATAAGGCAGAC --3’
R,5’- TTCAGGGCAGGTGGTAAGC --3’
F, 5’-TACCTTTGCCTTTGTCTGCC --3’
F, 5’-TAATCTGGGTGCCGTTGC --3’
F, 5’-ATCAAGGAACAGAAGCAACG --3’
F, 5’- GGAAGGTGGGCAGAGTAACA --3’ a
F, forward; R, reverse.
The PCR cycle consisted of AmpliTaq Gold activation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 58°C for 1 min. A dissociation curve was generated at the end of each PCR cycle to verify that a single product was amplified using software
provided with the 7000 Sequence Detection System. The changes in fluorescence of SYBR Green I dye in each cycle were monitored by the system software, and the calculated threshold cycle ( C t
) for each gene amplification was normalized to C t
of the 18S rRNA gene amplified from the corresponding sample before calculating the fold change from a selected time point to 0 time point using the following formula: fold change = 2
– C t where C t
for gene j = ( C t, j
– C t,18S rRNA
) a time point
– ( C t, j
– C t,18S rRNA
) time point 0
.
The real-time RT-PCR assays results were showed in
Table 2. The relative fold change for 10 genes listed in Table 1 determined by quantitative real-time RT-PCR and microarray hybridization results.
0 hr
ESTs Cluster
4 hr 10hr 15r
R a M b R a M b R a M b R a M b
DW679821 I 1 1 0.458 0.4827956 0.393 0.3528672 0.325 0.3337636
DW694001
DW698978
DW691154
DW699406
DW699957
DW702524
DW683219
DW699255
IV
IV
III
III
V
II
II
V
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1
1.648
3.817
7.571
3.331617
2.8971517
2.4231938
0.858 1.223877
16.851 3.6974249
1.472
2.65
2.183
1.1364942
1.65402885
3.1367872
1.516
4.082
17.239
1.877
9.624
1.327
4.125
1.452
4.7668537
3.1678551
18.971541
2.0662205
3.0518661
1.3442048
1.90301182
1.8660252
2.66
3.287
18.647
1.495
9.515
2.008
2.396
1.823
5.7810314
2.9819751
22.226249
2.4574132
3.6493771
1.6315185
1.981882
1.8301019 r
DW700354 V 1 1 4.304 4.1661408 3.135 2.7891405 4.093 2.5168176 0.879 a
Column R is the fold change relative to time point 0 hr determined by quantitative real-time RT-PCR; b C olumn M is the fold change relative to time point 0 hr determined by microarray hybridization results.
The data in Table2 showed that there was a strong positive correlation (r> 0.79) between the two techniques.
0.996
0.859
0.979
0.962
0.807
0.892
0.898
0.792
0.93




