AUTOCOVARIANCE STRUCTURES FOR RADIAL AVERAGES IN SMALL ANGLE X-RAY SCATTERING EXPERIMENTS
advertisement

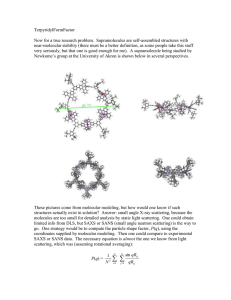
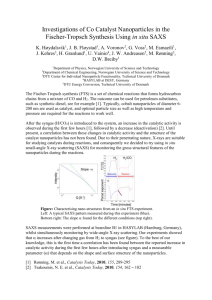
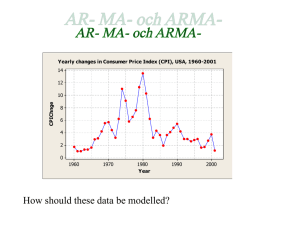
AUTOCOVARIANCE STRUCTURES FOR RADIAL AVERAGES IN SMALL ANGLE X-RAY SCATTERING EXPERIMENTS Andreea Erciulescu, F. Jay Breidt, Mark van der Woerd Departments of Statistics and Biochemistry & Molecular Biology, Colorado State University, Fort Collins Abstract Remove the mean to study autocorrelation Small-angle X-ray scattering (SAXS) is a relatively simple experimental method to obtain low-resolution molecular information, using high-intensity X-rays. In analyzing the recorded SAXS data, it is common to find data quality issues, such as detector saturation, low signal-to-noise ratio, radiation damage to the sample and adverse effects from sample concentration. In current analysis methods these issues are found after the experiment is complete, too late for adjustments in experimental protocols. Developing a rigorous statistical methodology for immediate assessment of data quality in raw SAXS images, without preprocessing, requires estimation of the autocovariance structure of the errors in the images and their radial averages. Autocorrelation results are stable Asymptotic Bartlett Bounds: ±1.96 ÷ √ m, where m = 32 in our case. We would expect exceedances of about 5%. SAXS experiments and images The investigator typically exposes different concentrations of the molecule in solution at different exposure times, collecting digital images of scattering patterns for each combination of concentration and exposure time. The images are subsequently reduced to a one- Consider the difference in replicate images to subtract the mean Problems: heteroskedasticity and residual mean structure Are local sample autocorrelations significantly different from zero? dimensional curve, which is interpreted and ultimately used for particle reconstruction. Nearest neighbors show autocorrelation Generating a SAXS Image φ e tter a c S -ray tX iden Inc kI kS ay -r dX 2θ kI Theoretical model q De te ct Sample autocorrelations are consistent with ”kernel convolution” model: RR 1 s RR 1 s I(q) = G( τ )µ(q + s)ds + G( τ )σ(q + s)Z(q + s)ds τ2 τ2 or Sample where Z(q) is an iid random field with mean 0 and variance 1. This has a physical interpretation due to detector engineering. Data processing Conclusion Neigboring pixels in the image data are in fact correlated, while farther removed pixels are uncorrelated. Future Directions Locally-estimated autocorrelations appear small, globally homogeneous, and isotropic (not direction-dependent). 1D Intensity profile Fourier Transform Particle shape reconstruction Consider a range of experimental conditions • Investigate model properties • Incorporate globally-estimated autocorrelations into statistical tests. The importance of autocorrelation Samples 2. Protein only (Glucose Isomerase) • We need to know confidence intervals on intensity data and derived data interpretation, ultimately for molecular shape reconstructions 1. Buffer (solvent) • Investigate various experimental conditions with new data sets. • Ensure best data quality possible for molecular envelope reconstructions. • Appropriately track uncertainty throughout the process. Instruments 3. DNA only • Current interpretation methods assume Gaussian error model and independent pixels • We apply correlation analysis to our two-dimensional images to test if individual pixels in the images are uncorrelated Two different instruments 4. Both protein and DNA Acknowledgements This research was supported in part by Award #R01GM096192 from the Joint NSF/NIGMS Initiative to Support Research in the Area of Mathematical Biology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of General Medical Sciences or the National Institutes of Health. Bibliography Brockwell, P.J. and Davis, R.A. (1991). Time Series: Theory and Methods, 2nd ed. Springer-Verlag, New York.