Multiple Covariates and More Complicated §16.4) Designs in ANCOVA (
advertisement
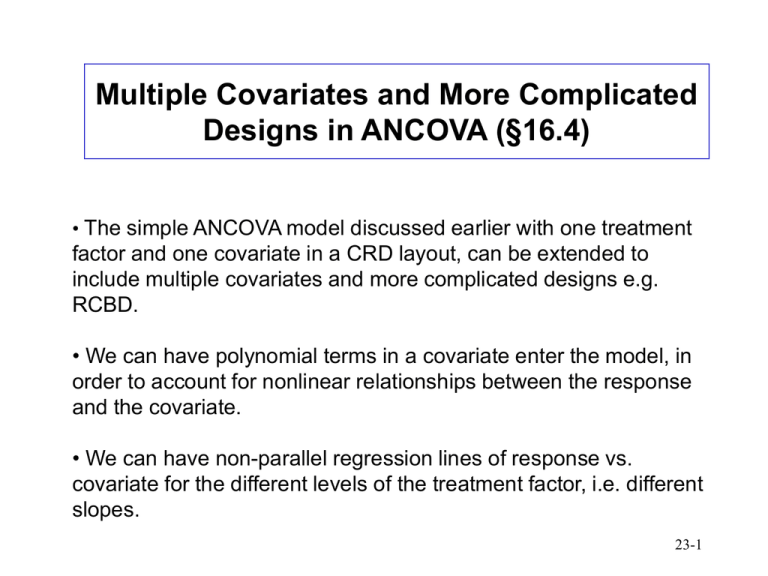
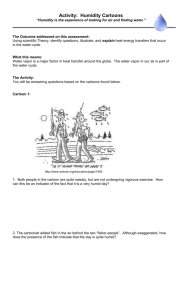
Multiple Covariates and More Complicated Designs in ANCOVA (§16.4) • The simple ANCOVA model discussed earlier with one treatment factor and one covariate in a CRD layout, can be extended to include multiple covariates and more complicated designs e.g. RCBD. • We can have polynomial terms in a covariate enter the model, in order to account for nonlinear relationships between the response and the covariate. • We can have non-parallel regression lines of response vs. covariate for the different levels of the treatment factor, i.e. different slopes. 23-1 Ex: Evaluation of Cool-Season Grasses for Putting Greens (§16.5) • Examine performance of three cultivars of turfgrass for use on golf course putting greens. These are resistant to diseases that are of concern to greenskeepers. Treatments: cultivars (C1,C2,C3). • Performance measure of interest is the speed the ball travels at on the green; measured by recording distance travelled after being rolled onto the green from a fixed height at a fixed angle. The farther the ball rolls, the faster the green. Response: speed (feet). • Eight regions of the country were selected for study (among them FL and AZ). Each region had a golf course with 3 putting greens available, and the cultivars were randomly assigned to the greens. Blocks: regions (R1,…,R8). • Thus factors affecting speed associated with geographic location were controlled through blocking. The only factor affecting speed the researchers were not able to control for was humidity. Thus this was recorded and used as a covariate. Covariate: humidity (% relative humidity). • Result: RCBD with 8 blocks, 3 treatments, and a single covariate (24 obs). 23-2 Linear relation with humidity plausible, but possibly non-parallel lines. 23-3 Linear Model yijk 0 i j j xijk ijk 0 = intercept of regression of y (speed) on x (humidity). i = effect of block (region) i=1,…,b (b=8). j = effect of treatment (cultivar) j=1,…,t (t=3). j = slope of regression of y on x for treatment j Index k=1,…,n denotes the replication (n=1). In our case we have a total of b*t*n=8*3*1=24 obs. 23-4 Formulate as Regression Model: Define Variables Y = speed X1 = humidity X2 = indicator of region 2 (1 if R2, 0 otherwise) . . . X8 = indicator of region 8 (1 if R8, 0 otherwise) X9 = indicator of cultivar 2 (1 if C2, 0 otherwise) X10 = indicator of cultivar 3 (1 if C3, 0 otherwise) 23-5 Model 1: treatment and block differences with covariate having unequal slopes • Allows for region and cultivar differences. • Allows cultivars within a region to have different slopes, but assumes a given cultivar slope is the same across regions. (Humidity can have unequal slopes.) Y 0 1 X 1 8 X 8 9 X 9 10 X 10 11 X 9 X 1 12 X 10 X 1 Interaction between humidity and cultivar allows for different cultivar slopes 23-6 Model 2: treatment and block differences with covariate having equal slopes • Allows for region and cultivar differences. • Cultivars have same slopes. (Humidity has equal slopes.) Y 0 1 X 1 8 X 8 9 X 9 10 X 10 23-7 Model 3: block but no treatment differences with covariate having equal slopes • Allows for region differences, but no cultivar differences. • Cultivars have same slopes. (Humidity has equal slopes.) Y 0 1 X 1 8 X 8 23-8 Compare Model 1 to Model 2: Should covariate (humidity) have equal slopes for each of the treatments (cultivars)? > model1 <- lm(speed ~ humid + region + cult + humid*cult) > model2 <- lm(speed ~ humid + region + cult) > model3 <- lm(speed ~ humid + region) # compare models 1 to 2 (test significance of different cult slopes) > anova(model2,model1) Analysis of Variance Table Model 2: Model 1: Res.Df 1 13 2 11 speed ~ humid + region + cult speed ~ humid + region + cult + humid * cult RSS Df Sum of Sq F Pr(>F) 0.47127 0.31203 2 0.15923 2.8067 0.1035 Conclude Model 2 is better (explain…) 23-9 Compare Model 2 to Model 3: Are there treatment (cultivar) differences? # compare models 2 to 3 (test significance of cult) > anova(model3,model2) Analysis of Variance Table Model 3: speed ~ humid + region Model 2: speed ~ humid + region + cult Res.Df RSS Df Sum of Sq F Pr(>F) 1 15 14.5661 2 13 0.4713 2 14.0948 194.41 2.063e-10 *** Conclude Model 2 is better (explain…) 23-10 Final Model (Model 2) There are treatment (cultivar) and block (region) differences, with equal covariate (humidity) slopes # final model > anova(model2) Analysis of Variance Table Response: speed Df Sum Sq Mean Sq F value Pr(>F) humid 1 3.0786 3.0786 84.9233 4.604e-07 *** region 7 1.2418 0.1774 4.8937 0.006737 ** cult 2 14.0948 7.0474 194.4050 2.063e-10 *** Residuals 13 0.4713 0.0363 23-11 Model 2 (Fitted Coefficients) > summary(model2) Call: lm(formula = speed ~ humid + region + cult) Coefficients: Estimate (Intercept) 8.421762 humid -0.022765 region2 -0.072989 region3 -0.084832 region4 -0.186642 region5 0.434006 region6 0.340397 region7 0.433041 region8 0.252458 cult2 0.917971 cult3 1.885567 Std. Error t value Pr(>|t|) 0.169847 49.584 3.34e-16 *** 0.002453 -9.281 4.25e-07 *** 0.155935 -0.468 0.6475 0.155846 -0.544 0.5954 0.168924 -1.105 0.2892 0.164553 2.637 0.0205 * 0.158506 2.148 0.0512 . 0.164995 2.625 0.0210 * 0.155974 1.619 0.1295 0.095581 9.604 2.87e-07 *** 0.095644 19.714 4.55e-11 *** 23-12 Compute adjusted cultivar means; Tukey comparisons # average region (block) effect mregion <- sum(c(-0.072989,-0.084832,0.186642,0.340397,0.434006,0.433041,0.252458))/8 # adjusted (for region and humid) means of C1, C2, C3 muC1 <- 8.421762-0.022765*mean(humid)+mregion muC2 <- 8.421762+0.917971-0.022765*mean(humid)+mregion muC3 <- 8.421762+1.885567-0.022765*mean(humid)+mregion # Tukey multiple comparisons > library(multcomp) > summary(glht(model2, linfct=mcp(cult="Tukey"))) Multiple Comparisons of Means: Tukey Contrasts Fit: lm(formula = speed ~ humid + region + cult) Linear Hypotheses: Estimate Std. Error t value p value 2 - 1 == 0 0.91797 0.09558 9.604 <1e-07 *** 3 - 1 == 0 1.88557 0.09564 19.714 <1e-07 *** 3 - 2 == 0 0.96760 0.09520 10.164 <1e-07 *** 23-13 Plot humidity vs. cultivars; check fit # Check model fit pdf(file="Plots/golf3.pdf",pointsize=3,width=6,height=5) par(mfrow=c(2,1)) qqnorm(model2$res) plot(model2$fitted,model2$res); abline(0,0) # plot speed vs. humid (add fitted cult lines) plot(humid,speed,type="n",xlab="Humidity",ylab="Speed",main=" Speed vs. Humidity With Fitted Cultivar Lines") text(humid,speed,as.character(cult)) abline(8.421762,-0.022765,lty=1) # cult 1 abline(8.421762+0.917971,-0.022765,lty=2) # cult 2 abline(8.421762+1.885567,-0.022765,lty=3) # cult 3 legend(80,9.8,c("Cultivar 1","Cultivar 2","Cultivar 3"),lty=c(1,2,3)) 23-14 23-15 23-16