
Using the Results of Genetic Studies (Genomics)
Using the results of genetic studies Over the last few sessions you have been learning about some of the experimental techniques and analyses that allow us to determine how our genetics link to phenotypes and diseases. As our knowledge of the genome and other -omics have increased, so has the complexity of the results that we obtain. In this session, we will be focusing on some of the challenges that can occur when trying to interpret these datasets and, using examples of real genome-wide studies, will examine how clinically and biologically useful information can be extracted. You will learn about how GWAS and RNAseq data can complement each other, enhancing our ability to link genetics and transcriptomics to specific phenotypes. We will also look at how to validate the results of these studies. Some of the techniques described in this session involve advanced and complex statistical analyses. You are not expected to understand these, but should be able to discuss the ideas underpinning these experimental designs. Learning Outcomes On successful completion of the lecture, students should be able to: Discuss the difficulties in interpreting and analysing data from genome-wide experiments such as GWAS Understand how GWAS results are linked to causal genes via fine mapping and analysis of SNP locations Discuss how RNAseq data can be used to annotate GWAS data via eQTL mapping Discuss how to use in vitro and in vivo experiments to validate results from genome-wide studies
-
What is the importance of human genetic disorders in studying body systems? (2)
Unique Window: Human genetic disorders provide a unique insight into the regulation of normal body systems and how genetic mutations influence physiological processes.
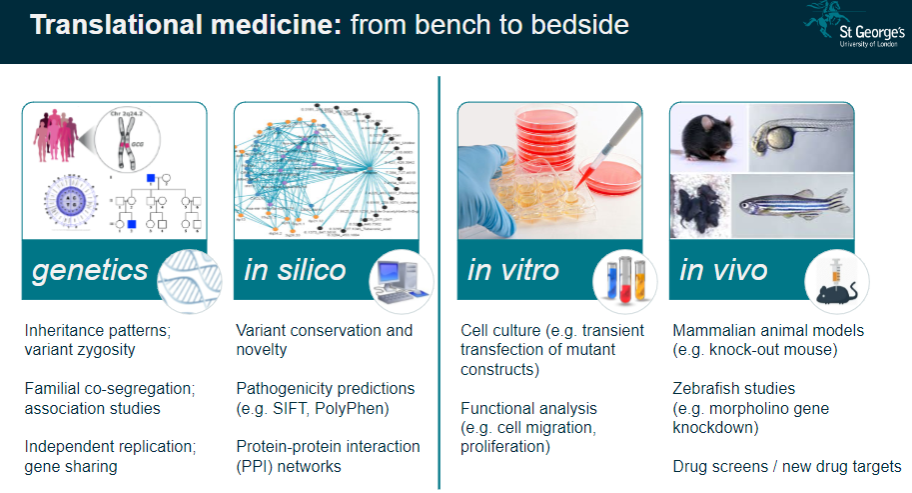
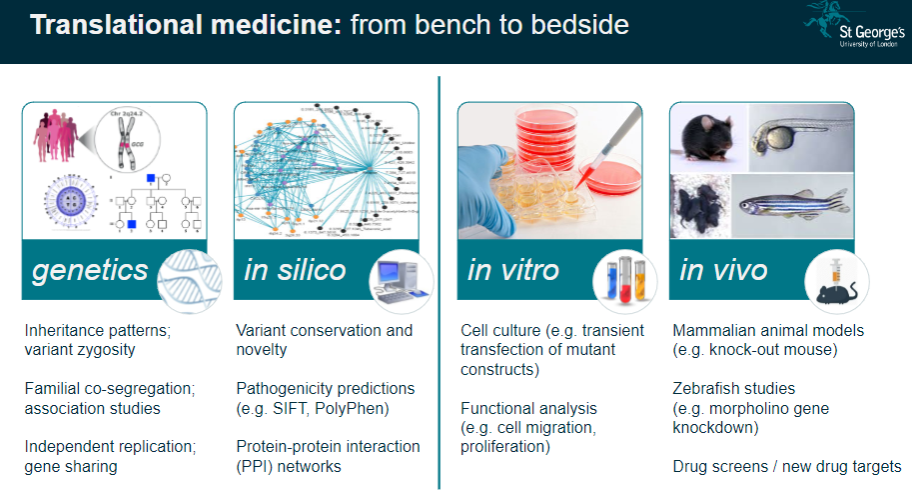
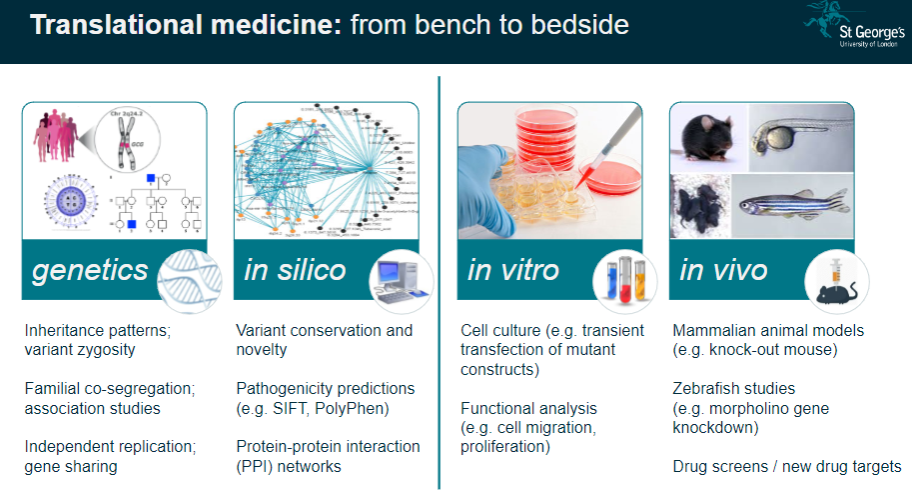
Translational Medicine: These studies help translate genetic findings from the lab (bench) to clinical applications (bedside), improving disease understanding and treatment.
-
What are the challenges in interpreting genetic study results? (3)
Complexity: Genetic data often involve large numbers of variants, making it challenging to pinpoint the exact cause of a condition.
Pathway Analysis: Understanding how identified genes interact in biological pathways can be difficult due to the complexity of gene networks.
Individual Variability: Results can vary across individuals, requiring careful consideration of personal genetic backgrounds and environmental factors.
-
What is pathway analysis in genomics? (2)
Understanding Pathways: Pathway analysis examines how specific genetic variants affect biological processes or pathways in the body.
Link to Disease: This approach helps to link genetic findings with the development of diseases by understanding how genes interact in functional pathways.
-
How is personalized genetics used in genomics? (2)
Individual Genetic Makeup: Personalized genetics involves using an individual's unique genetic profile to guide healthcare decisions and treatments.
Tailored Medicine: It aims to customize treatment plans based on genetic predispositions, ensuring more effective and individualized medical care.
-
What is Expression Quantitative Trait Loci (eQTL) mapping? (3)
Definition: eQTL mapping identifies genetic variants that influence gene expression levels in specific tissues.
Gene-Expression Link: It helps link genetic variation to changes in gene activity, which can influence disease susceptibility.
Analysis: Researchers can analyze the effects of genetic variants on gene expression in order to better understand complex diseases.
-
What is co-localisation analysis in genomics? (2)
Definition: Co-localisation analysis identifies whether genetic variants that influence gene expression also affect disease traits.
Purpose: This analysis helps determine whether a variant contributes to disease risk through its effects on gene expression.
-
How does genetic variation contribute to disease pathogenesis? (3)
Genomic Hits to Pathology: Genetic variants, also known as genomic 'hits,' can directly influence the development of diseases by altering normal cellular processes.
Gene Identification: Understanding which genes are involved in disease allows for targeted therapies.
Cell-Specific Impact: Some genetic variants may only affect specific cell types, which is important when studying disease mechanisms.
-
What is functional phenotyping in genomics? (3)
In vitro Validation: This involves studying genetic mutations in cultured cells to observe their effects on cell behavior, such as migration or proliferation.
In vivo Validation: Animal models, like knockout mice, are used to validate genetic findings and study the physiological impacts of mutations.
Reverse Genetic Screens: These are used to investigate the effects of specific gene mutations by systematically altering genes and observing the resulting phenotypes.
-
What role do mammalian animal models (e.g., knockout mice) play in genomics? (3)
Knockout Mice: These animals are genetically modified to lack specific genes, allowing researchers to study the effects of these gene deletions on development and disease.
Modelling Disease: Knockout mice serve as models for human diseases, providing valuable insight into genetic mutations and their physiological consequences.
Therapeutic Testing: Animal models are crucial for testing potential treatments before clinical trials in humans.
-
How are zebrafish used in genomics research? (2)
Morpholino Gene Knockdown: Zebrafish are used to study gene function by knocking down gene expression using morpholinos.
Disease Modelling: Zebrafish models are useful for studying developmental diseases and testing new therapeutic approaches.
-
What is the role of drug screens in genomics? (2)
New Drug Targets: Drug screens are used to identify potential drug targets based on genetic findings, aiming to find effective treatments for diseases.
Target Validation: These screens can help confirm whether a particular gene or pathway is a viable target for drug development.
-
What is the significance of variant conservation and novelty in genetic studies? (3)
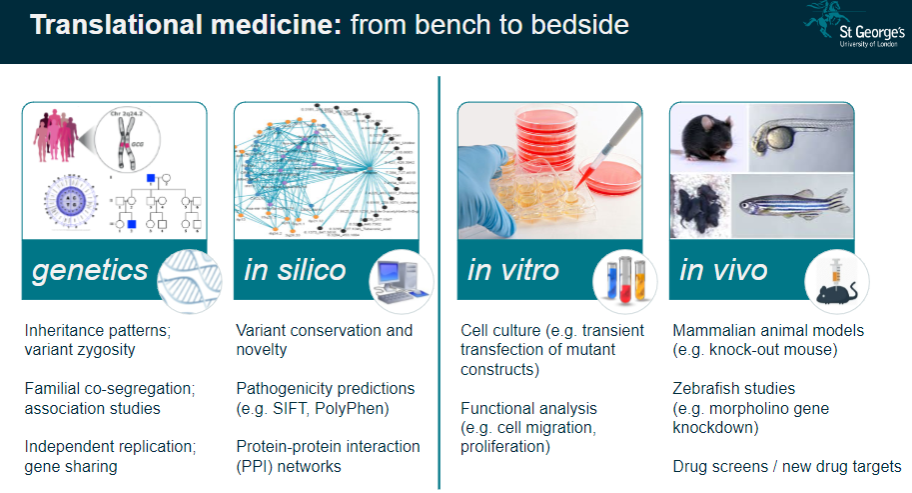
Variant Conservation: The conservation of a genetic variant across species can indicate its functional importance and potential relevance to disease.
Novelty: Identifying novel variants that have not been previously associated with diseases can lead to new discoveries in genetics.
Pathogenicity Predictions: Tools like SIFT and PolyPhen predict whether a variant is likely to be pathogenic based on its conservation and function.
-
Picture demonstrating Genetic analysis pipelines:
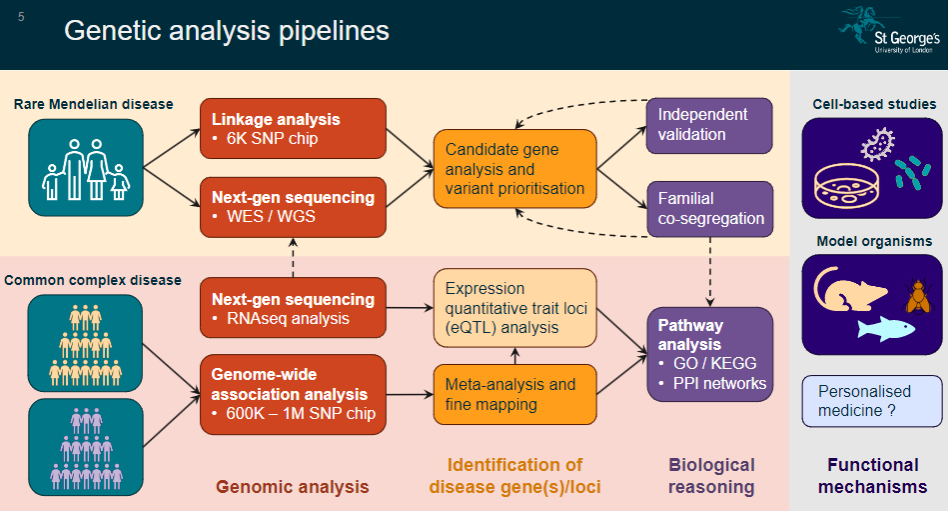
-
What defines a 'rare' disease? (1)
A disease that affects <1 in 2,000 people.
-
What are the challenges in genetic analysis of rare diseases? (3)
Genetic heterogeneity
Small patient populations
Lack of resources
-
What approaches are used in genetic analysis of rare diseases? (3)
Whole Exome Sequencing (WES)
Whole Genome Sequencing (WGS)
Gene panel testing
-
How does genetic heterogeneity impact the diagnosis of rare diseases? (2)
Multiple causes for the disease
Hard to identify mutations
-
How does a small patient population impact genetic analysis of rare diseases? (2)
Less data for analysis
Hard to validate findings
-
How does a lack of resources challenge genetic analysis of rare diseases? (2)
Fewer genetic databases
More research needed
-
What is genetic heterogeneity in the context of rare disease genomics? (1)
It refers to the presence of different genetic causes of the same disease, meaning the disease can result from mutations in different genes or genomic regions.
-
What is locus heterogeneity and how does it affect rare disease analysis? (2)
Locus heterogeneity occurs when different subsets of families with the same disease show linkage to different regions of the genome.
This complicates the identification of a specific disease-causing region across various families.
-
What is clinical heterogeneity and how does it impact rare disease diagnosis? (2)
Clinical heterogeneity refers to variability in how the disease manifests in patients, leading to different clinical presentations of the same condition.
It can result in misdiagnosis as the disease may appear similar to other conditions with different underlying causes.
-
What is incomplete penetrance in rare disease genomics? (2)
Incomplete penetrance occurs when not everyone with the disease-causing variant expresses the disease traits.
Some individuals may remain asymptomatic carriers despite carrying the mutation.
-
How does insufficient data coverage affect linkage analysis in rare disease genomics? (2)
Smaller pedigrees (family studies with fewer members) lack the statistical power to generate a statistically significant LOD score.
This leads to many peaks of "suggestive" linkage without definitive evidence.
-
Why is genetic marker distribution important in rare disease linkage analysis? (2)
Genetic markers may not be uniformly distributed across the genome, resulting in large gaps between them.
This causes large linkage intervals, complicating the identification of precise disease-causing regions.
-
What is locus heterogeneity in rare diseases? (2)
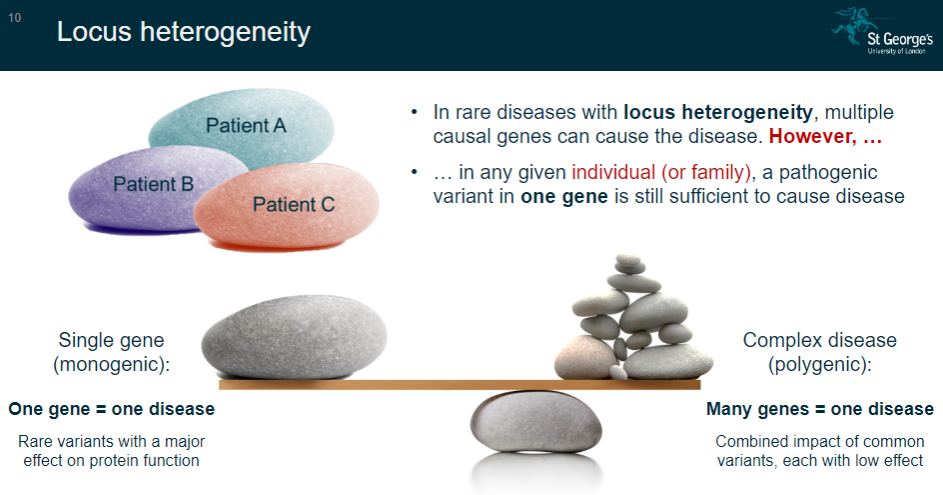
Locus heterogeneity occurs when multiple causal genes can lead to the same disease.
However, in any given individual or family, a pathogenic variant in one gene is still enough to cause the disease.
-
What is the concept of single-gene (monogenic) diseases? (2)
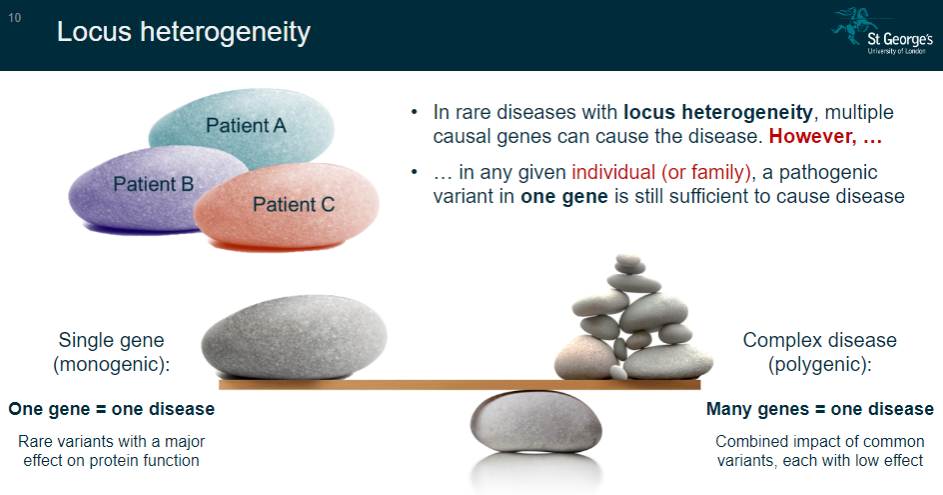
In single-gene diseases, one gene is responsible for causing the disease.
These diseases are often caused by rare variants that have a major effect on protein function.
-
What is the concept of complex (polygenic) diseases? (2)
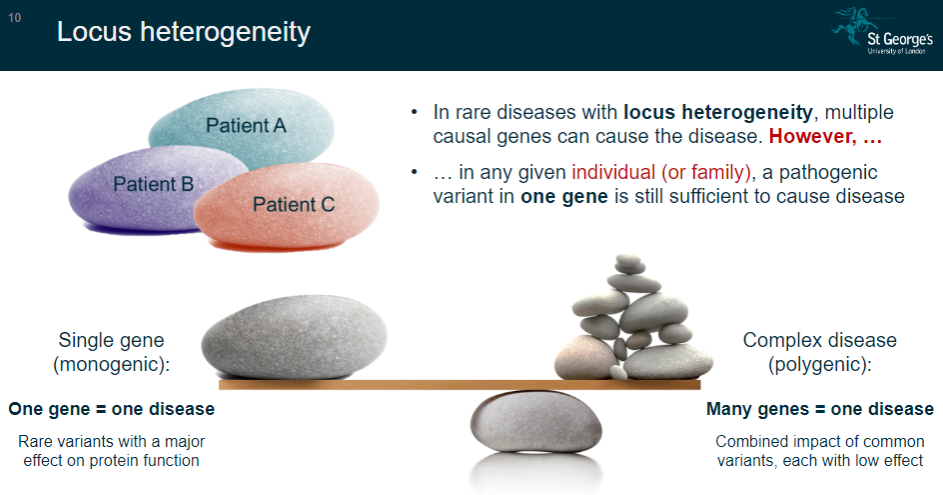
Complex diseases involve many genes contributing to the disease.
These diseases are influenced by the combined impact of common variants, each with a low effect on the disease outcome.
-
How does incomplete penetrance affect linkage analysis? (2)
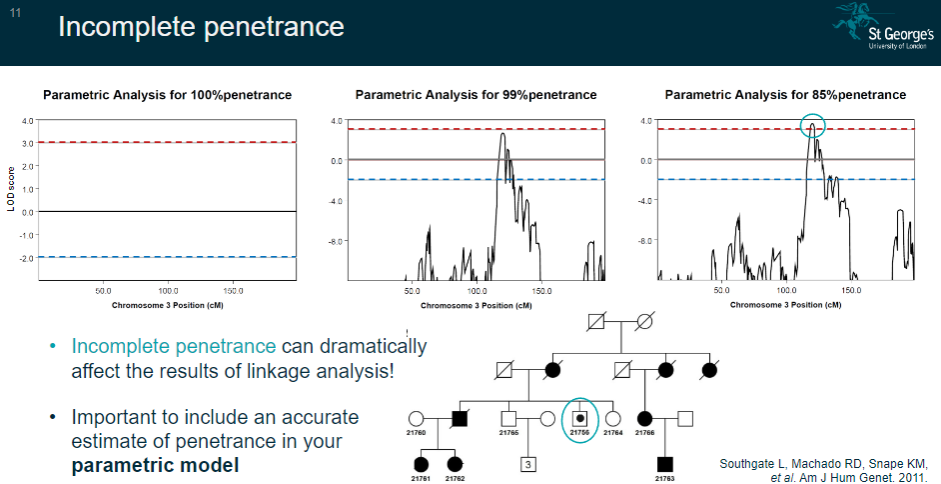
Incomplete penetrance can significantly impact the results of linkage analysis by introducing uncertainty in the expression of the disease.
It’s crucial to include an accurate estimate of penetrance in the parametric model to account for these effects.
-
How does data coverage in small pedigrees affect linkage analysis, and how can it be improved? (3)
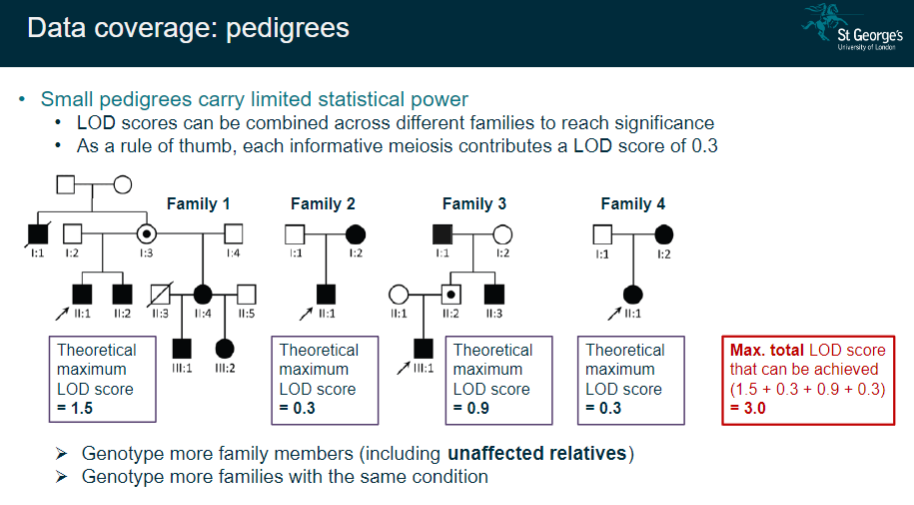
Small pedigrees have limited statistical power, which can affect the accuracy of the results.
LOD scores from different families can be combined to increase the statistical significance.
To improve data coverage, it is helpful to genotype more family members (including unaffected relatives) and increase the number of families with the same condition.
-
How are SNP and microsatellite markers used in linkage analysis? (3)
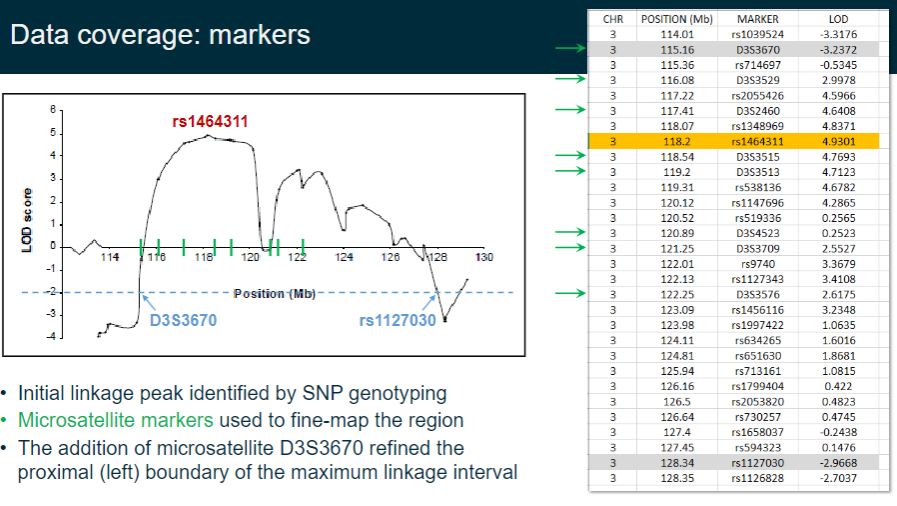
Initial linkage peaks are identified using SNP genotyping.
Microsatellite markers are then used to fine-map the region.
The addition of microsatellite D3S3670 helps refine the proximal (left) boundary of the maximum linkage interval.
-
Picture demonstrating Challenges in rare disease genomics: NGS:

-
Picture demonstrating Genetic counselling:

-
How can identifying a rare disease gene help in understanding biological pathways? (3)
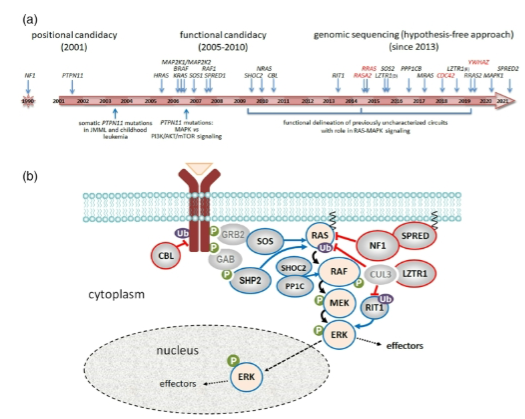
Identification of a rare disease gene reveals disrupted biological pathways.
Disrupting other genes or proteins within the same pathway may lead to the same phenotype.
This can help in developing new drug targets or repositioning existing drugs for treatment.
-
Picture demonstrating Rare disease pathways as potential drug targets:
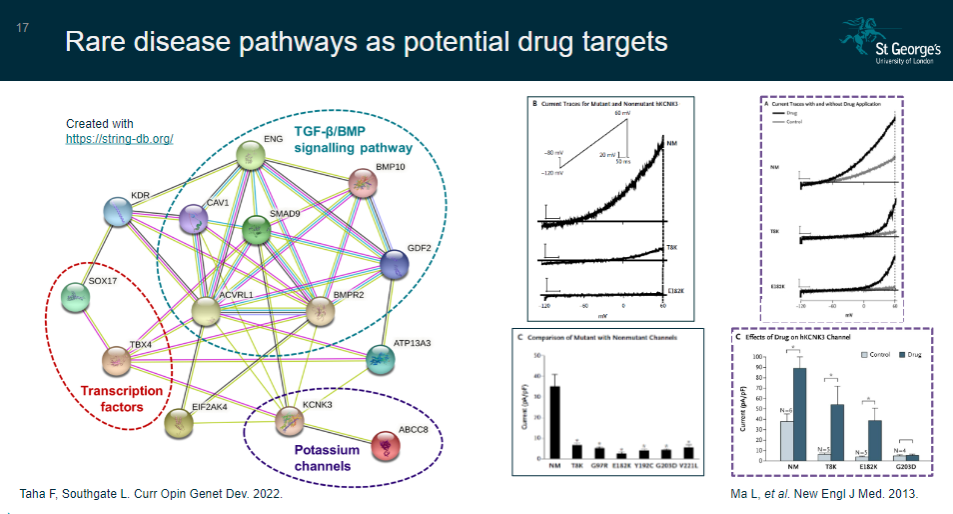
-
Picture demonstrating The promise of gene therapy:
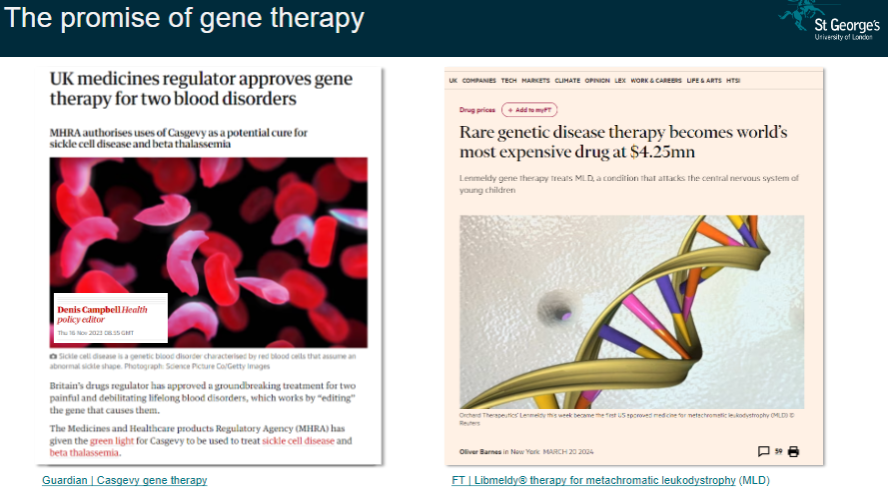
-
Picture demonstrating Genetic screening and drug development:
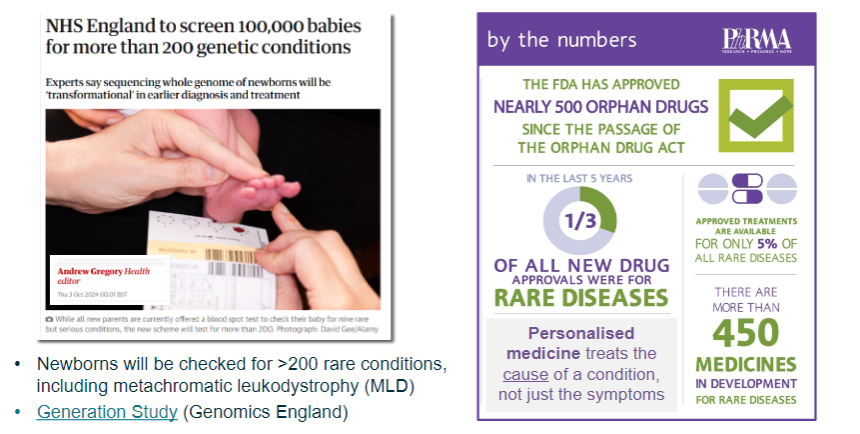
-
Picture demonstrating Genetics of common complex disease:
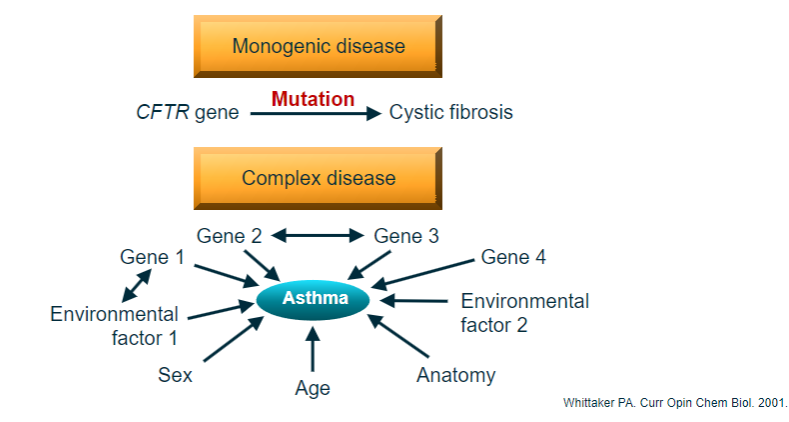
-
What is the difference between monogenic and polygenic diseases? (3)

Monogenic (Single gene): One gene causes one disease, typically due to rare variants with a major effect on protein function.
Polygenic (Complex disease): Many genes contribute to one disease, where the combined impact of common variants, each with a low effect, leads to disease.
Polygenic diseases: Require multiple ‘hits’ in different genes or loci, but the combinations of genetic hits vary across individuals.
-
Why is studying common diseases important? (5)
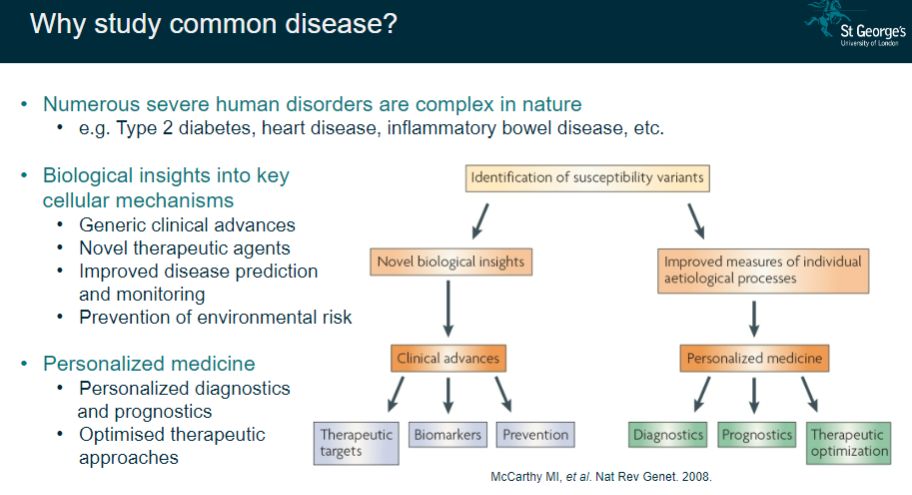
Numerous severe disorders: Common diseases like Type 2 diabetes, heart disease, and inflammatory bowel disease are complex and widespread.
Biological insights: Provides a deeper understanding of key cellular mechanisms.
Clinical advances: Leads to general improvements in clinical practice.
Therapeutic innovations: Helps in developing novel therapeutic agents and personalized medicine.
Improved disease management: Allows for better prediction, monitoring, prevention, and optimized therapeutic approaches.
-
What is personalized medicine, and why is it important? (4)
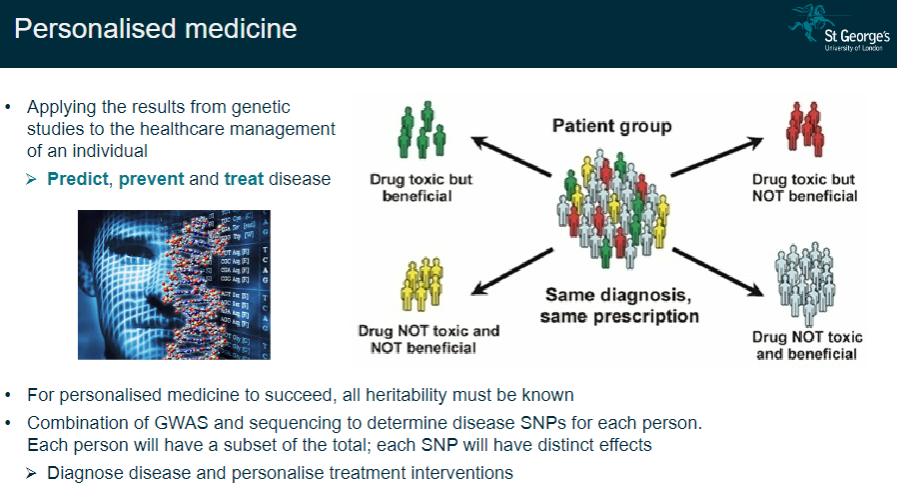
Complete heritability knowledge: All heritable factors must be understood for personalized medicine to work effectively.
GWAS + sequencing: Combining Genome-Wide Association Studies (GWAS) and sequencing to identify disease-related SNPs specific to each individual.
Personalized treatment: Use genetic data to diagnose diseases and tailor treatments for each person.
Predict, prevent, and treat: Genetic information allows for prediction, prevention, and more effective treatment of diseases.
-
What are the challenges after obtaining GWAS results in common disease genomics? (5)
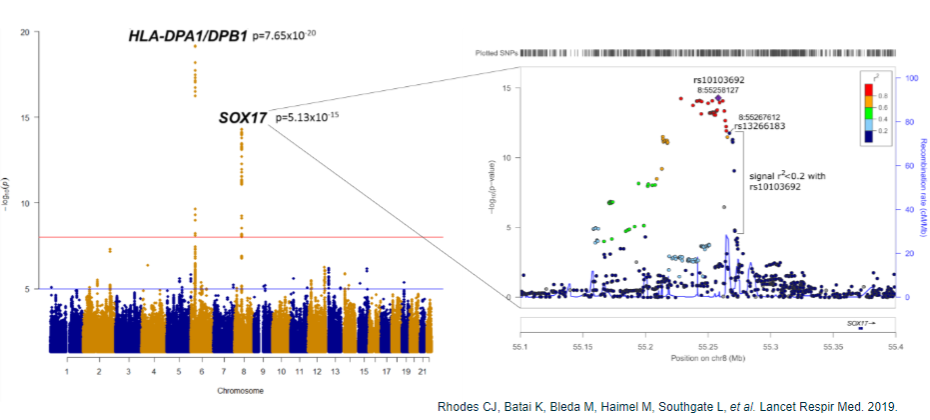
Population-specific associations: Associations may be specific to certain populations, requiring validation in other populations.
Multiple loci: Large GWAS often reveal thousands of association peaks, making prioritization of findings difficult.
Non-coding SNPs: 90% of associated SNPs are located in non-coding regions, complicating the identification of causal genes.
Identifying causal variants: Determining which variants are actually causal remains challenging.
Mechanism of action: Understanding the molecular mechanism and identifying the relevant tissue or cell type involved in the disease process.
-
Picture demonstrating Independent validation of GWAS hits:
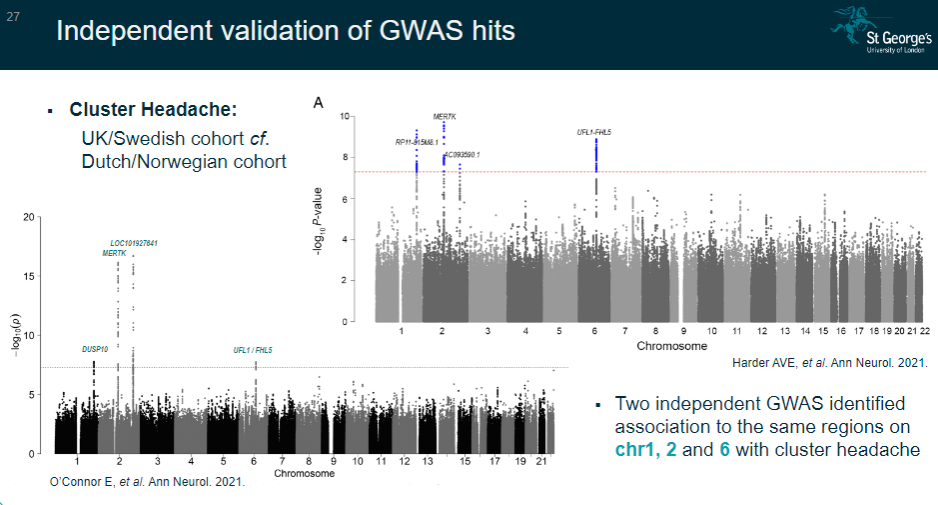
-
Picture demonstrating Meta-analysis of the combined cohort:
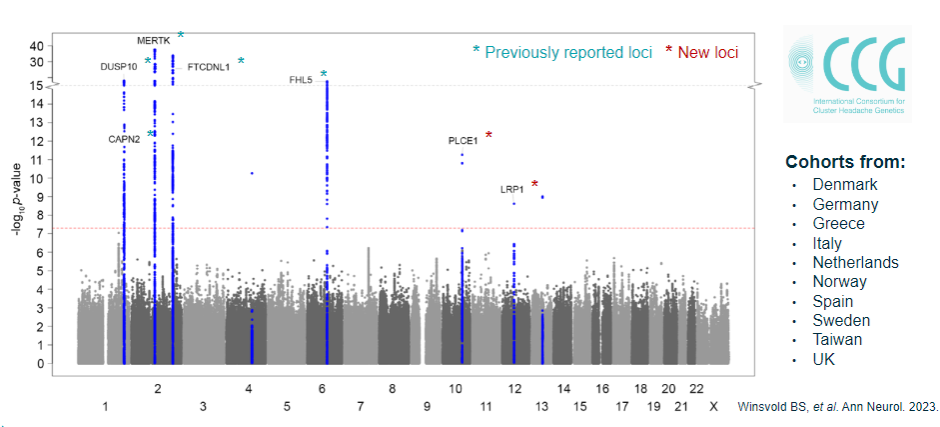
-
How do you interpret multiple GWAS loci detected in complex diseases? (4)
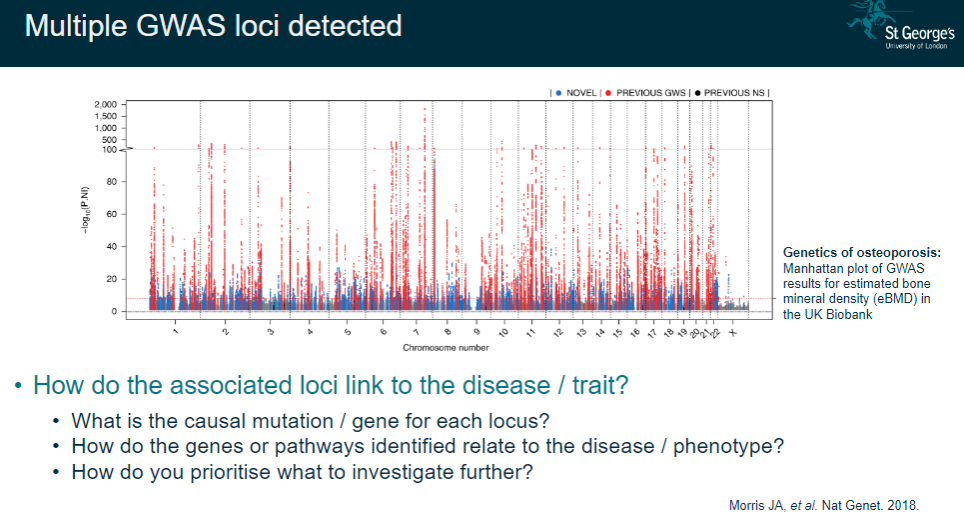
Link to disease/trait: Understanding how the loci identified contribute to the disease or phenotype.
Causal mutation/gene: Identifying the specific mutation or gene responsible for each associated locus.
Gene/pathway relationship: Investigating how the genes and pathways identified are involved in the disease process.
Prioritization: Deciding which loci or genes to focus on for further investigation, based on their relevance to the disease.
-
What is fine mapping in GWAS and how is it performed? (4)
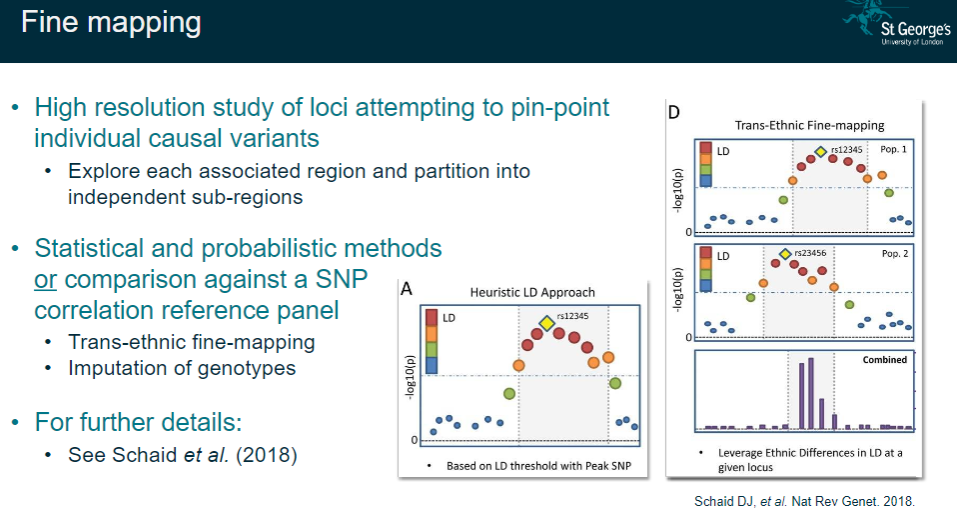
High-resolution study: Aiming to pinpoint individual causal variants within a locus.
Sub-region partitioning: Exploring each associated region and dividing it into independent sub-regions for further analysis.
Statistical/probabilistic methods: Using advanced techniques or comparison with SNP correlation reference panels to refine the regions of interest.
Trans-ethnic fine-mapping: Performing fine-mapping across different populations and imputing genotypes for more accurate results.
-
What are the challenges in linking GWAS loci to genes? (5)
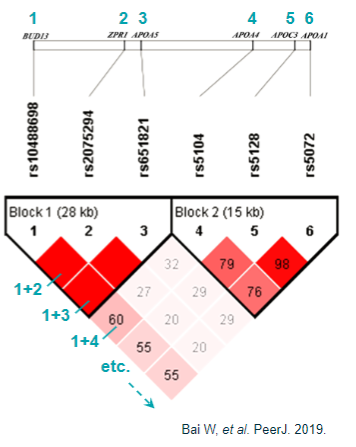
Linkage disequilibrium (LD): Makes it challenging to distinguish the causal variant due to LD's influence by multiple factors beyond recombination.
Haplotype blocks: May not always provide reliable fine-mapping of complex traits because they can be influenced by various factors.
Non-coding SNPs: 90% of GWAS SNPs are located in non-coding or intergenic regions, complicating the identification of causal genes.
Regulatory elements: SNPs might be located in regulatory regions (e.g., promoters, enhancers, transcription factor binding sites), not directly in coding regions.
Long-range effects: The lead SNP may affect genes at a distance via mechanisms like Topologically Associating Domains (TADs), which complicates pinpointing the affected gene(s).
-
Picture demonstrating Which is the causal gene..?:
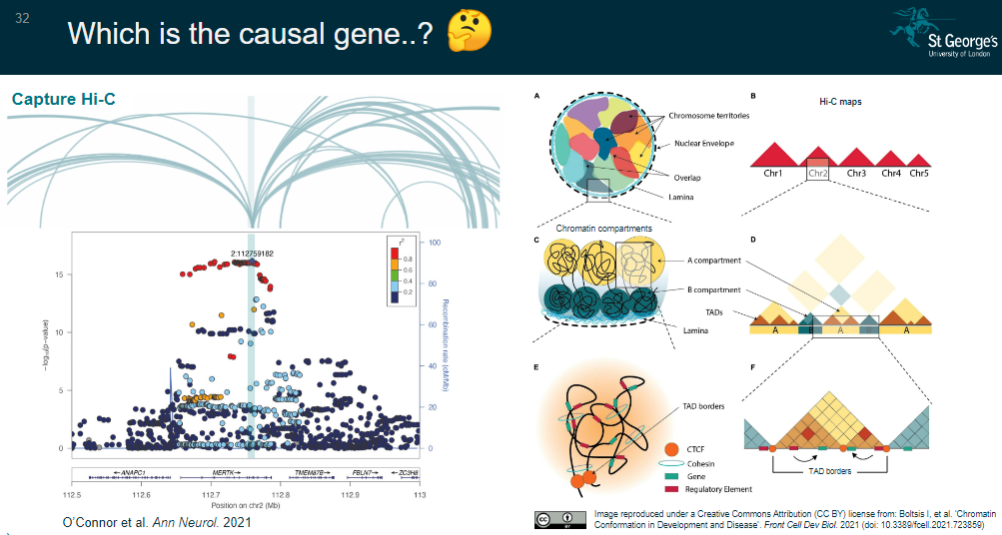
-
How are target causal genes assigned? (5)
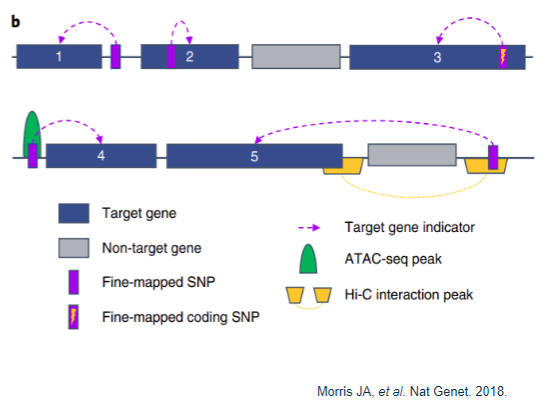
Proximity:
Gene closest to a fine-mapped SNP.
Gene containing a fine-mapped SNP.
Non-synonymous exonic change:
Fine-mapped SNP is a coding variant that results in an amino acid change.
Chromatin conformation capture:
Gene closest to a fine-mapped SNP residing in an ATAC-seq peak.
Fine-mapped SNP located in a Hi-C promoter interaction peak, indicating it is closer to the target gene in 3D space than linearly on the genome.
-
What are the key steps in translating GWAS results to biological reasoning? (4)
Identify new biology: Understand what the loci reveal about biological processes.
Determine pathogenesis: Explore how the findings explain disease mechanisms.
Assess biological or clinical importance: Identify findings relevant to diagnosis, treatment, or drug targets.
Validate findings: Confirm results through experimental or clinical studies.
-
What are the main features of GWAS in studying common complex diseases? (6)

Identifies genomic loci associated with a phenotype/disease.
Detects SNP associations across the entire genome.
Reveals a large number of loci.
Does not identify causal variants or genes.
Fails to pinpoint cell type, tissue, or developmental stage.
-
What are the main features of RNA sequencing (RNA-seq) in gene expression studies? (6)
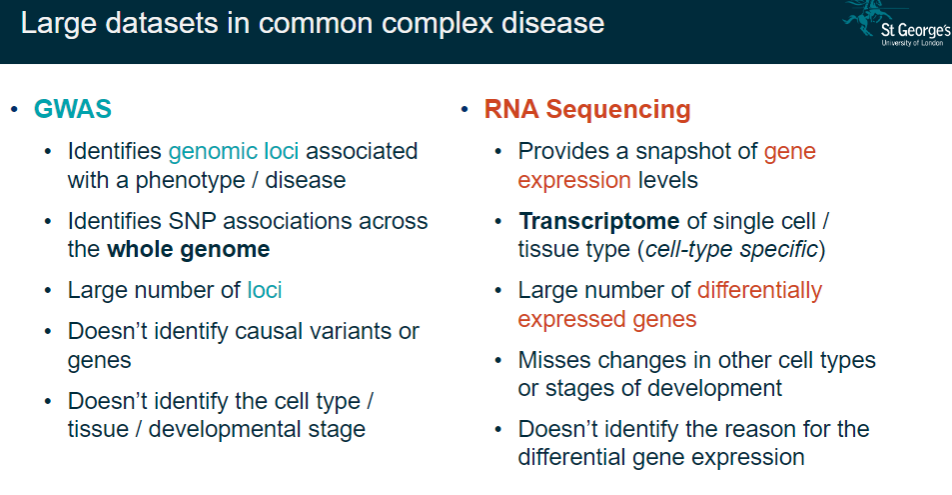
Provides a snapshot of gene expression levels.
Captures the transcriptome of single cells or specific tissues.
Identifies a large number of differentially expressed genes.
Misses changes in other cell types or stages of development.
Does not identify the reasons for differential expression.
-
What does RNA-seq data provide, and how is it analysed? (6)

Relative expression data for every gene.
Identifies differentially expressed genes (up-regulated in red; down-regulated in blue).
Requires setting significance thresholds (e.g., p-value, fold change).
May reveal novel allele-specific expression and alternative transcripts.
Importance of using a relevant tissue/cell type.
-
What is single-cell RNA sequencing (scRNA-seq), and what does it reveal? (2)
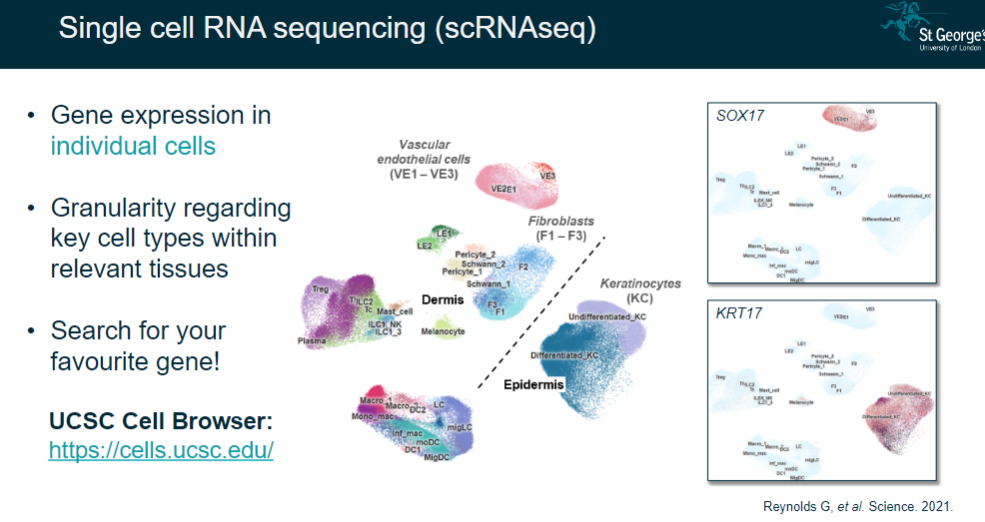
Measures gene expression in individual cells.
Provides granularity regarding key cell types within relevant tissues.
-
What are the challenges in genomic analysis using RNA-seq? (6)
Too Many Results: RNA-seq often detects numerous changes in gene expression, making it hard to separate meaningful data from technical noise.
Snapshot Limitation: The transcriptome reflects gene expression at a single moment in time, which may not fully capture dynamic changes.
Tissue Relevance: It's essential to study the right tissue or cell type related to the disease or condition, or the data might not be meaningful.
Time Sensitivity: Gene expression can vary over time, during different stages of development, or under disease conditions, complicating analysis.
Cell-Type Variation: Different cell populations within a sample can react differently to treatments, potentially obscuring results.
Validation Needed: While RNA-seq shows differential expression, it doesn't explain why these changes occur, requiring follow-up studies for confirmation.
-
What is pathway analysis in genomic studies? (3)

Generate a gene set and compare it to a previously annotated database.
Use resources such as Gene Ontology (GO) or Kyoto Encyclopaedia of Genes and Genomes (KEGG).
Identify which pathways or processes are enriched based on the gene set.
-
What is an expression quantitative trait locus (eQTL)? (2)
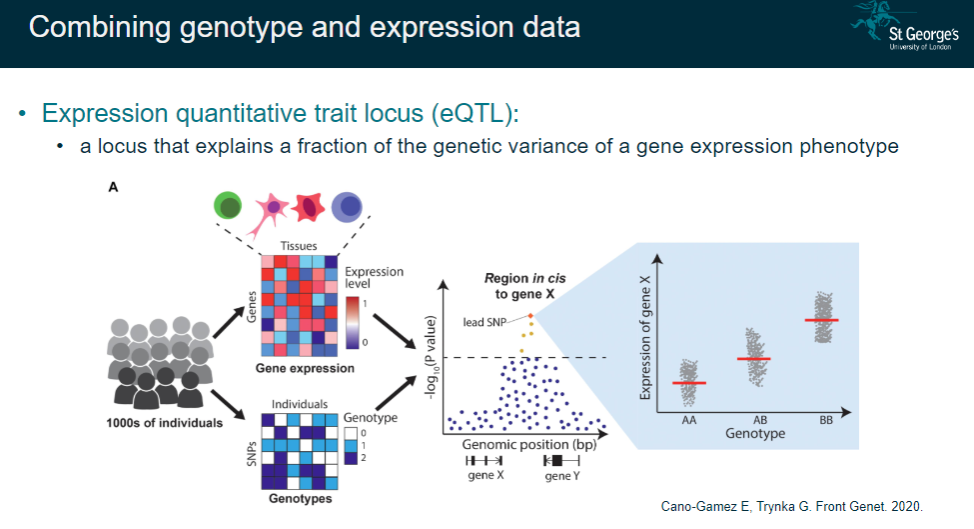
A locus that explains a fraction of the genetic variance in gene expression.
Combines genotype data with expression data to understand genetic regulation of expression.
-
What is co-localisation analysis and its purpose? (5)
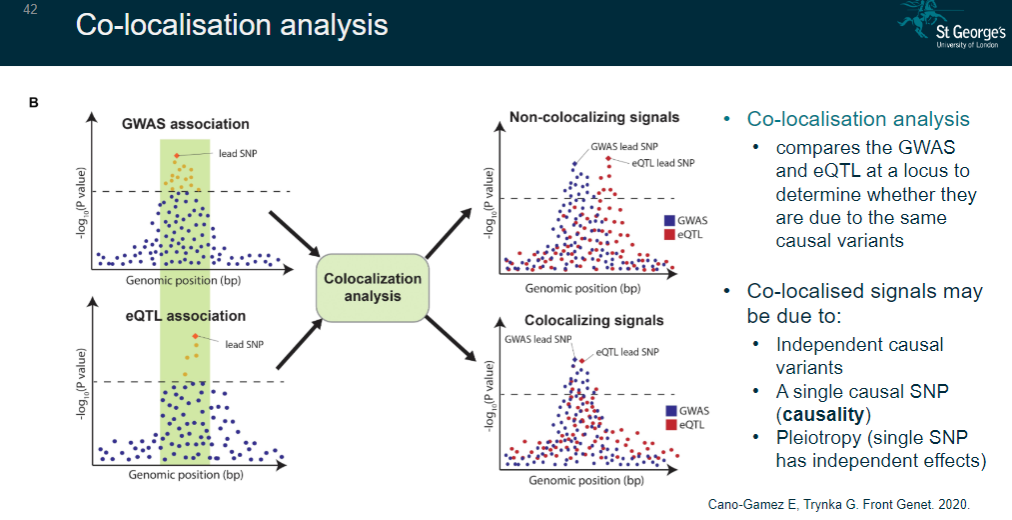
A method that compares GWAS and eQTL signals at a locus.
Determines whether they share the same causal variants.
Identifies whether co-localised signals are due to:
Independent causal variants.
A single causal SNP (causality).
Pleiotropy (a single SNP with independent effects).
-
Picture demonstrating Functional validation of candidate genes: examples:
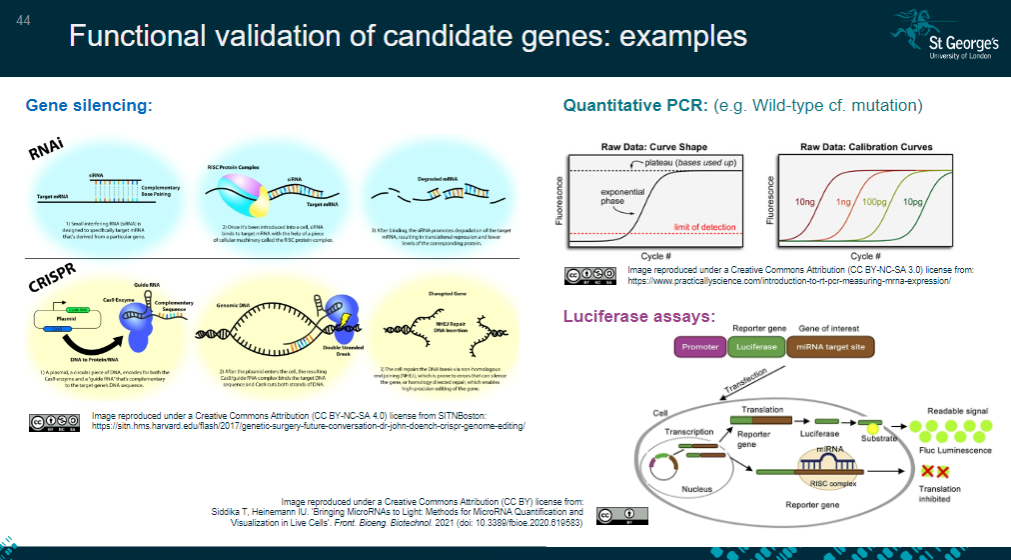
-
What is functional analysis in cell studies (in vitro)? (6)
Identifying the most relevant cell type for the study, often disease-specific.
Confirming gene/protein expression using techniques like qPCR or Western blot.
Determining protein localization within the cell via immunofluorescence studies.
Testing whether the variant impacts cell functionality using assays like luciferase.
Assessing cellular behaviors such as migration or proliferation.
Observing the cell's response to treatment or stimuli.
-
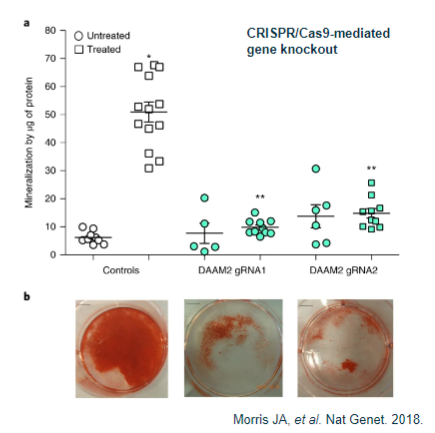
What is functional analysis in model animals (in vivo)? (2)
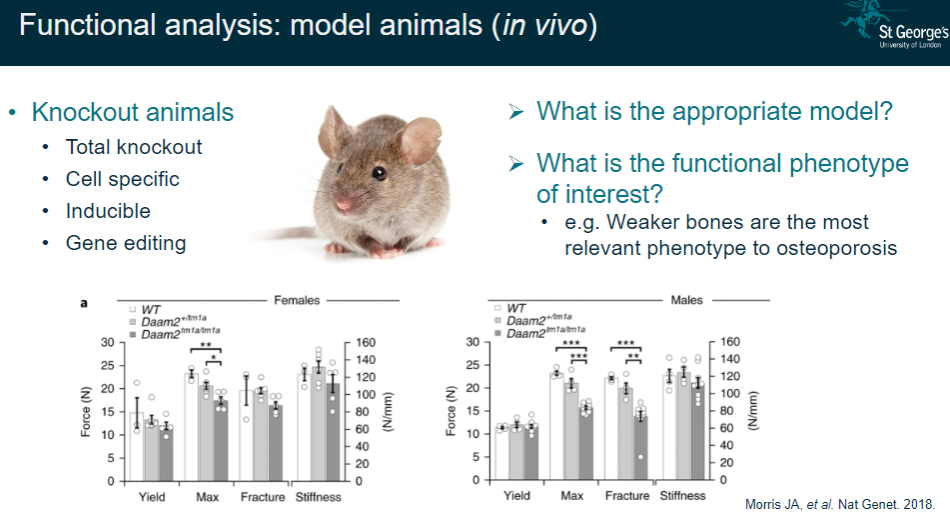
Determining the appropriate animal model for the study.
Identifying the functional phenotype of interest (e.g., weaker bones as a key phenotype in osteoporosis).
-
What are reverse genetic screens, and how do they differ from forward genetic screens? (2)
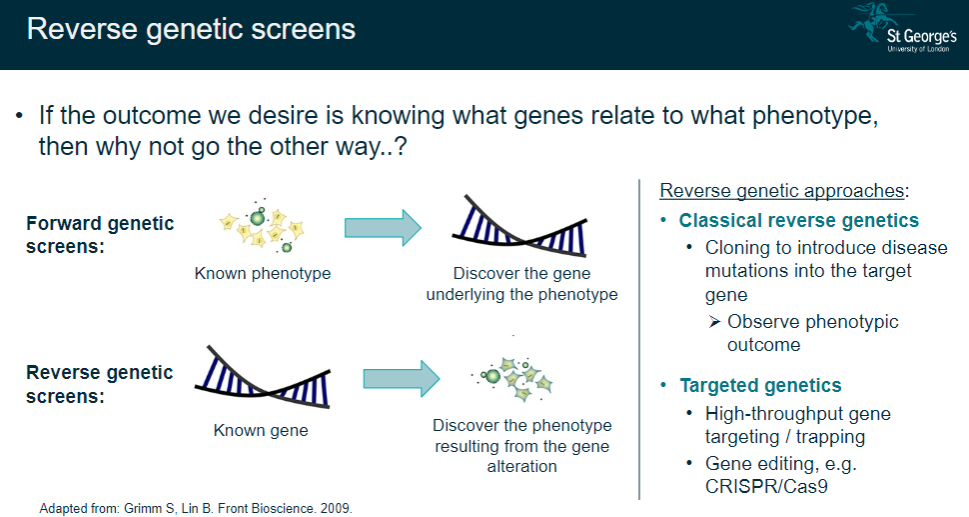
Reverse genetic screens start with a known gene to observe the resulting phenotype after alteration.
Forward genetic screens start with a known phenotype to discover the underlying gene.
-
What are the main approaches used in reverse genetics? (3)
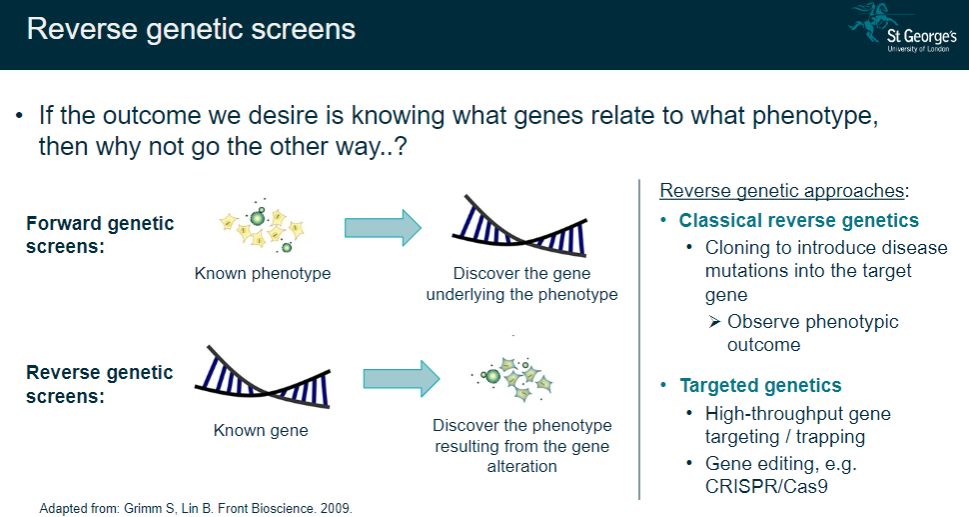
Classical reverse genetics: Introduce disease mutations into a target gene via cloning and observe the phenotypic outcome.
Targeted genetics: Use high-throughput gene targeting or trapping techniques.
Gene editing: Employ tools like CRISPR/Cas9 to edit specific genes and study the resulting phenotype.
-
What is the International Mouse Phenotyping Consortium (IMPC), and what is its goal? (2)
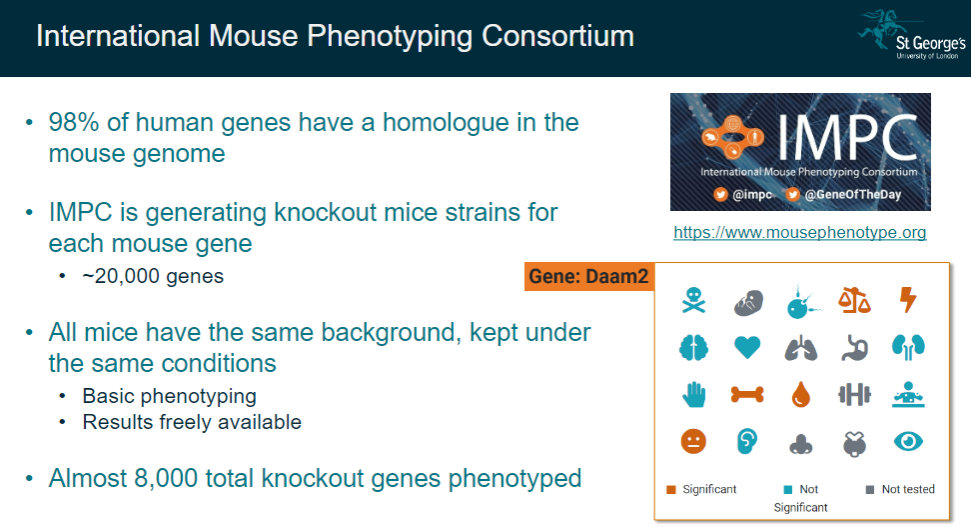
The IMPC aims to generate knockout mice strains for each mouse gene, as 98% of human genes have a homologue in the mouse genome.
The goal is to phenotype approximately 20,000 genes in mice to understand gene functions.
-
What are the key features of the IMPC's approach? (3)
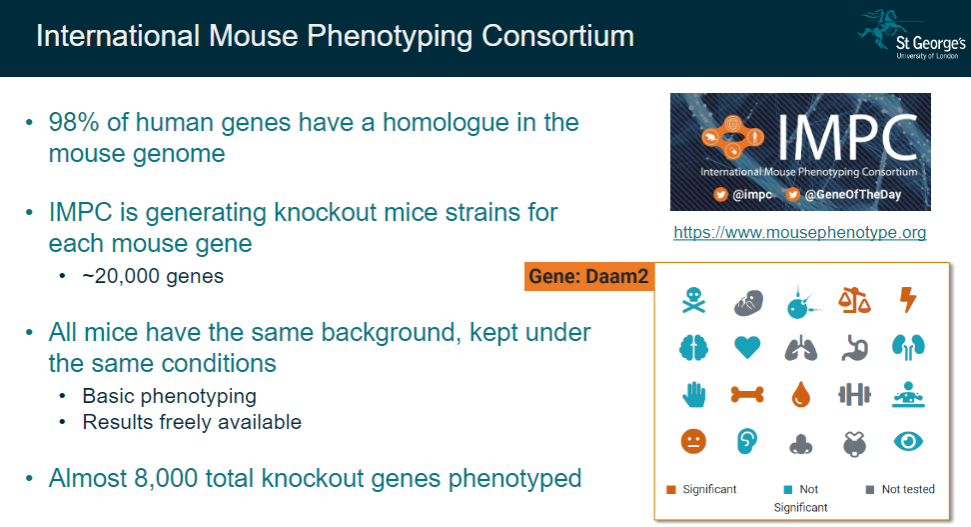
All mice used for the study have the same background and are kept under controlled conditions.
Basic phenotyping is performed to observe the impact of gene knockouts.
Results from these studies are made freely available, with almost 8,000 knockout genes already phenotyped.
-
What are the challenges in interpreting genome-wide genetic studies? (3)
Large datasets generated by genome-wide genetic studies can be difficult to interpret.
There are more loci than prioritization methodologies available.
Fine-mapping causal variants and linking the results to biological mechanisms can be complex.
-
How can the interpretation of genetic data be improved? (4)
Through RNA sequencing and eQTL analysis, which provide insights into gene expression.
By performing pathway analysis in silico.
Through functional validation using in vitro and in vivo methods.
Reverse genetic screens can offer hypothesis-free functional phenotyping data.

