
The Metagenome (Genomics)
SDL: The Metagenome The increasing throughput and ever decreasing costs of sequencing technologies are enabling the development of much more detailed characterisation of the microbiome. Novel sequencing platforms are changing the way we think about molecular research and diagnostics approaches to a range of different human and environmental problems. The aim of this session is to introduce the concept of the Metagenome. We will define the concept and then discuss methods available to understand and elucidate the microbiome and the metagenome in different biological contexts, including the more traditional approach of 16S rRNA targeted PCR amplification as well as more cutting edge approaches of shotgun metagenomic approaches. Learning Outcomes On successful completion of the lecture, students should be able to: Define the term metagenomics Define the term microbiome and be able to distinguish it from metagenomics Describe human microbiome environments Describe technological approaches to metagenomics, specifically targeted PCR amplification and whole genome shotgun sequencing Describe the reasoning behind choosing 16S RNA sequences for targeted amplification Describe why the kitome is an important consideration for metagenomics Session Resources Session recording:
-
What is metagenomics and why is it important? (3)
Metagenomics is the study of genetic material recovered directly from environmental or biological samples, such as soil, water, or the human body.
It allows for the analysis of microbial communities that are often difficult or impossible to culture in a lab.
It is important because it helps understand the genetic diversity of microbial communities and their roles in various ecosystems, including the human body.
-
What is the human microbiome and how is it related to metagenomics? (3)
The human microbiome refers to the collection of genomes of the microorganisms that live in and on the human body.
This includes bacteria, archaea, fungi, viruses, and other microbes.
Metagenomics is used to study the microbiome by analyzing the genetic material from these microorganisms directly, providing insights into their diversity, function, and interactions.
-
What are the key technological approaches in metagenomics? (4)
16S targeted amplification: Focuses on the 16S ribosomal RNA gene, which is commonly used to identify and classify bacteria in a sample.
Whole genome shotgun metagenomics: Involves sequencing all the DNA in a sample, allowing for the identification of all microorganisms present, not just bacteria.
Genomics: Studies the entire genome content of organisms in the sample.
Transcriptomics: Studies the gene expression (RNA content) of microorganisms in the sample.
-
What is the difference between genomics, transcriptomics, proteomics, and metabolomics? (4)
Genomics: The study of the entire genome sequence of an organism.
Transcriptomics: The study of gene expression by analyzing the RNA content (transcripts) of an organism.
Proteomics: The study of the entire protein content (proteome) of an organism.
Metabolomics: The study of the entire set of metabolites (small molecules) present in an organism, reflecting its metabolic state.
-
What is the significance of organisms not living in isolation? (2)
Organisms interact within complex mixtures of species in their environment, forming ecological communities.
Understanding these interactions is essential because most environments contain a diverse range of organisms, and studying them in isolation would not provide a complete picture.
-
What is the difference between microbiota and microbiome? (3)
Microbiota refers to the ecological community of microorganisms (bacteria, archaea, fungi, viruses, etc.) in a particular environment, such as the human body.
Microbiome refers to the collective genomes of these microorganisms and the "theatre of activity" where they interact within the ecosystem.
The microbiome encompasses the full genetic makeup and functional potential of the microbiota.
-
Why is metagenomics particularly useful in studying organisms that cannot be cultured? (2)
Many microorganisms cannot be cultured in laboratory settings, making traditional methods of studying them difficult.
Metagenomics allows researchers to bypass the need for culturing and study the genetic material of microorganisms directly from environmental samples.
-
What is the role of metagenomics in understanding complex microbial ecosystems? (3)
Metagenomics provides insights into the genetic diversity of microbial communities, which are often too complex to study with traditional methods.
It helps reveal the interactions between different microorganisms, including commensals and pathogens, within a shared environment.
By studying these ecosystems, metagenomics contributes to a deeper understanding of how microbial communities influence health, disease, and the environment.
-
Picture demonstrating the microbiome:
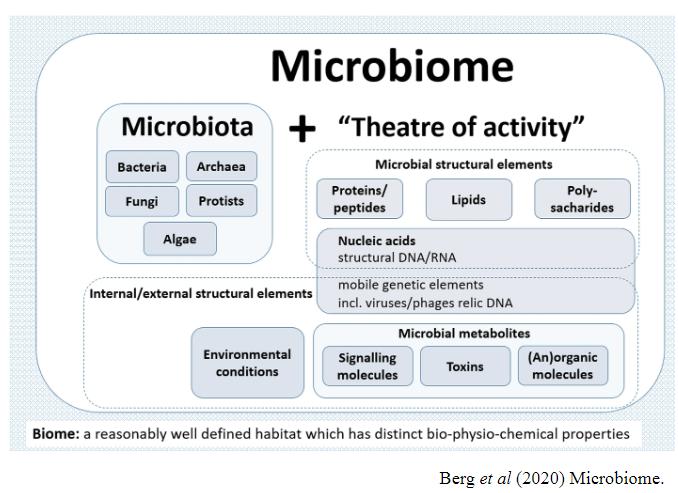
-
What are some examples of different microbiomes and their significance? (6)
Deep sea microbiome: Microorganisms in extreme ocean environments, important for biogeochemical cycles.
Soil microbiome: Microbes in soil that influence nutrient cycling and plant growth.
Hospital microbiome: Microbes in healthcare environments, impacting infection control and patient health.
Subway microbiome: Microorganisms in subway systems, relevant to public health and infection spread.
Gut microbiome: Microbes in the gastrointestinal tract, crucial for digestion, immune function, and health.
Skin microbiome: Microbes on the skin that protect against pathogens and regulate skin health.
-
What is the human microbiome and its role in health? (3)
The human microbiome is the collective genome of microorganisms living in and on the human body (e.g., gut, skin, mouth, etc.).
It is involved in critical processes such as immune regulation, digestion, and disease prevention.
Imbalances in the microbiome can lead to various health issues like obesity, autoimmune diseases, and skin conditions.
-
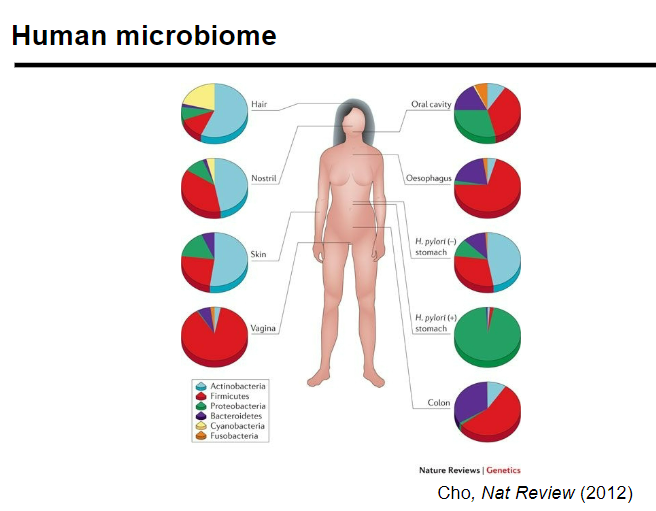
Picture demonstrating the human microbiome:
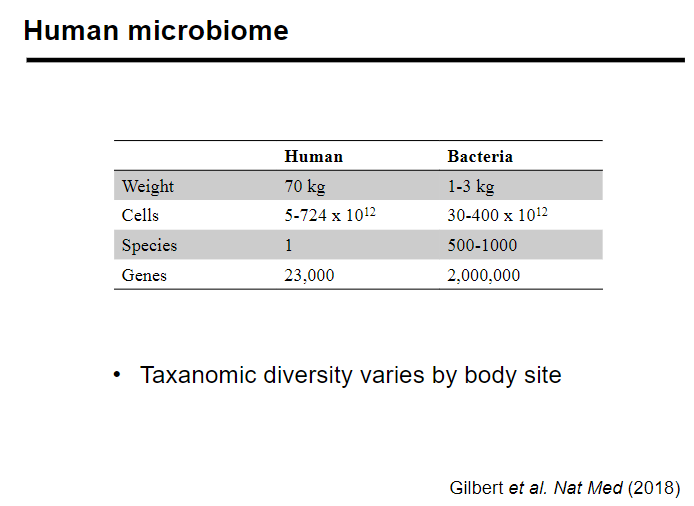
-
What makes the human microbiome unique to each individual? (2)
The human microbiome is unique to each person, with differences even between identical twins.
This uniqueness is influenced by a combination of genetics, environmental factors, and lifestyle.
-
What illnesses have been associated with changes in the microbiome? (4)
Irritable Bowel Syndrome (IBS)
Depression
Cancer
Other gastrointestinal and systemic diseases
-
How can the gut microbiome be used to classify individuals? (2)
The gut microbiome can classify individuals as lean or obese with over 90% accuracy.
This classification is based on microbial composition and its effects on metabolism.
-
What role does the early-life gut microbiome play in health? (2)
The early-life gut microbiome is linked to the development of allergic conditions, such as asthma.
Microbial imbalances during early development can influence immune system maturation.
-
What findings were observed in the microbiome during Clostridium difficile infection (CDI)? (3)
The stool microbiome during Clostridium difficile infection (CDI) differs significantly from that of healthy individuals.
CDI has a greater effect on the stool microbiome than the host's genetic factors.
Faecal microbiota transplantation (FMT) can cure CDI, rapidly restoring the stool microbiome to a healthy state.
-
What is the importance of gene content in metagenomics? (2)
In metagenomics, the gene content of microorganisms is often more important than the relative abundance of organisms.
This focus helps in understanding the functional potential of microbial communities, rather than just their composition.
-
What are the main technological approaches in metagenomics? (3)
Targeted PCR amplification: Focuses on specific genes for microbial identification and analysis.
16S rRNA sequencing: Used to analyze bacterial communities.
Internal Transcribed Spacer (ITS) sequencing: Used to study eukaryotic organisms, particularly fungi and certain protists.
Whole genome shotgun sequencing: Provides a comprehensive analysis of the entire genetic material of the community.
-
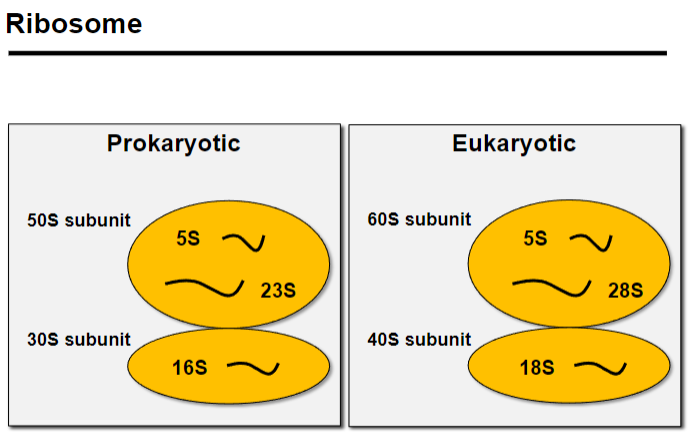
What is 16S targeted PCR amplification and its role in metagenomics? (3)
16S targeted PCR amplification is a method used to study bacterial communities by targeting the 16S ribosomal RNA gene.
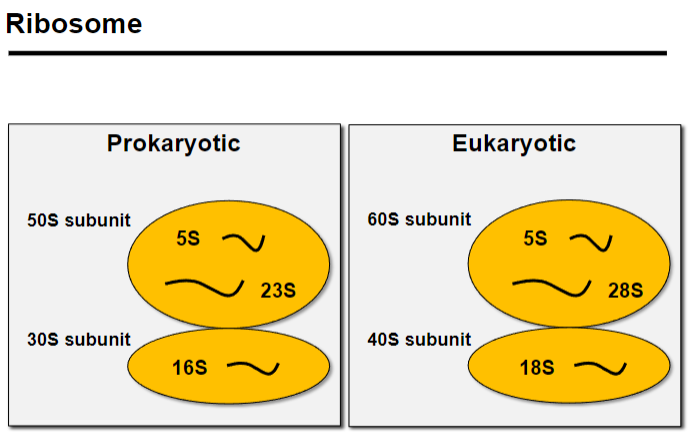
The 16S rRNA is a component of the 30S small subunit of the prokaryotic ribosome, essential for protein synthesis.
This method helps identify and classify bacteria based on conserved regions within the 16S gene.
-
What is 16S targeted PCR amplification and its workflow? (6)
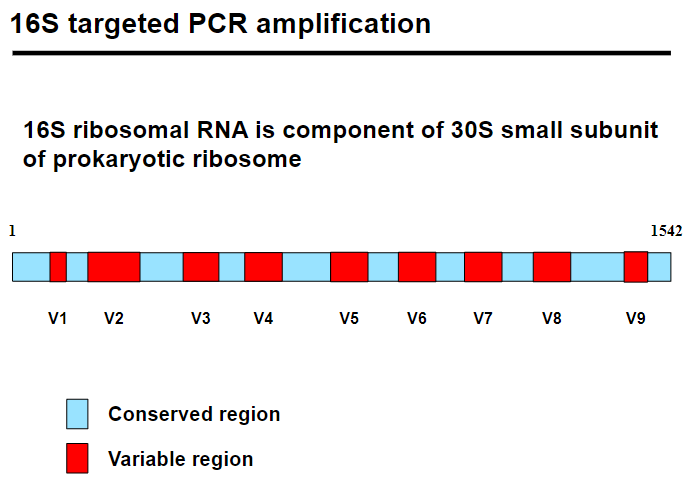
16S targeted PCR amplification studies bacterial communities by amplifying the 16S rRNA gene, part of the 30S small subunit of the prokaryotic ribosome.
The gene has conserved regions (identical across species) and variable regions (species-specific), enabling bacterial classification.
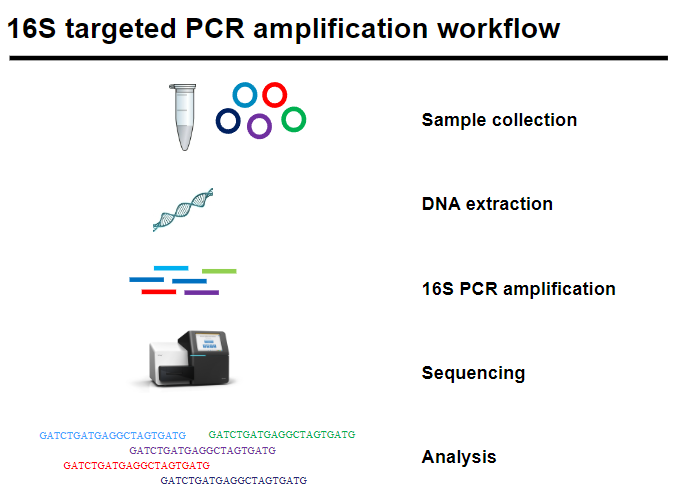
Workflow:
Sample collection: Obtain biological/environmental samples.
DNA extraction: Isolate DNA from samples.
16S PCR amplification: Amplify the 16S gene.
Sequencing: Sequence the amplified DNA.
Analysis: Analyze sequences to identify species and community composition.
-
What is an Operational Taxonomic Unit (OTU)? (3)
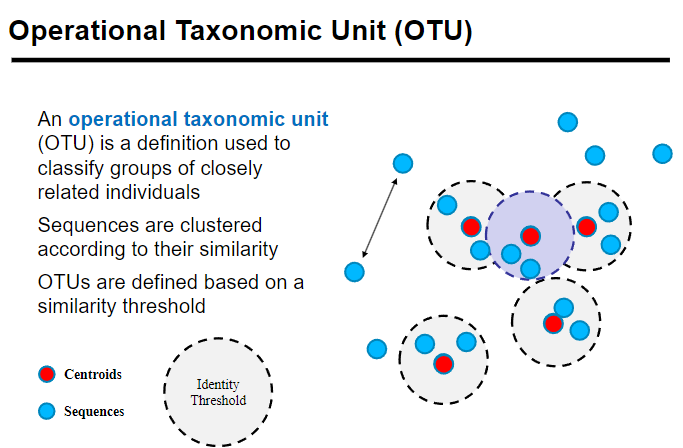
An Operational Taxonomic Unit (OTU) is used to classify groups of closely related individuals.
Sequences are clustered based on their similarity, grouping organisms with similar genetic sequences.
OTUs are defined by a similarity threshold, typically around 97% sequence identity, to determine species or genus-level grouping.
-
What is the 16S targeted PCR amplification workflow? (5)
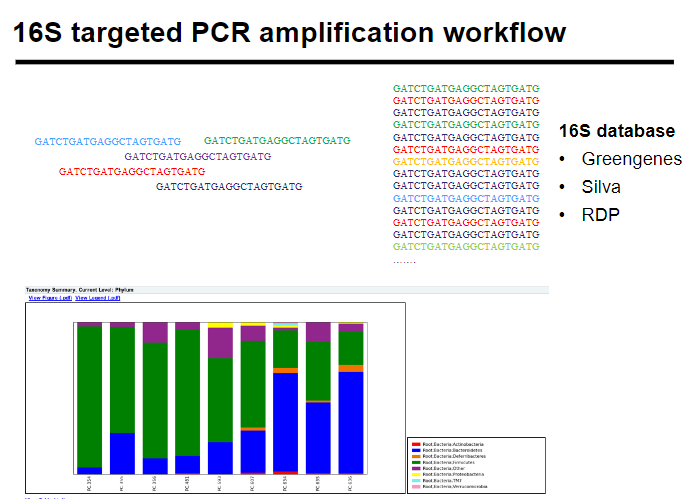
Sample collection: Collect biological/environmental samples.
DNA extraction: Isolate DNA from the samples.
16S PCR amplification: Amplify the 16S rRNA gene.
Sequencing: Sequence the amplified DNA.
Analysis: Analyze sequences to identify bacterial species and community composition.
-
What are common 16S databases? (3)
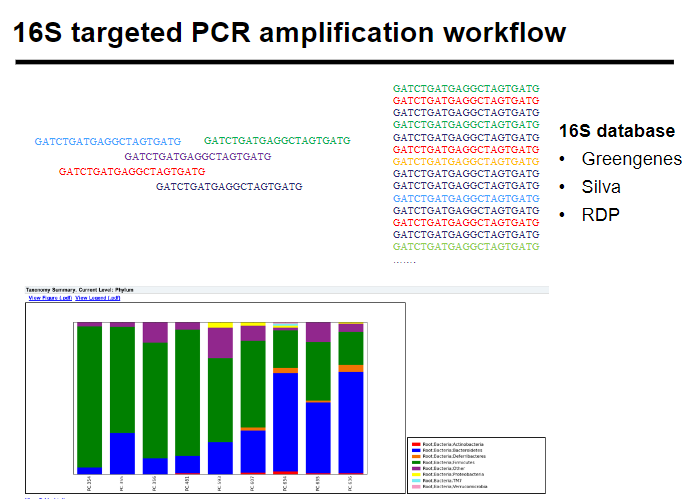
Greengenes: A curated 16S rRNA gene sequence database for phylogenetic classification.
Silva: A comprehensive database for ribosomal RNA sequences, especially useful in environmental microbiology.
RDP (Ribosomal Database Project): A resource for ribosomal RNA sequence-based analysis of microbial communities.
-
What are alpha and beta diversity in microbial ecosystems? (4)
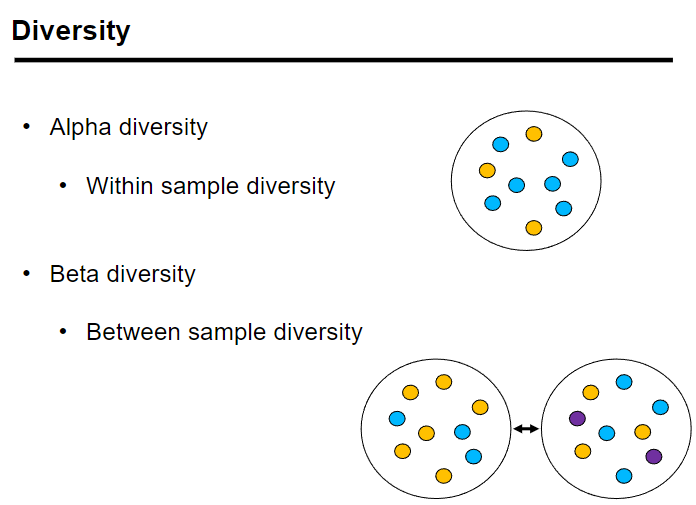
Alpha diversity: Measures the within-sample diversity, referring to the diversity of species within a single sample.
Beta diversity: Measures the between-sample diversity, comparing diversity between different samples.
Alpha diversity can be quantified by metrics like species richness and species diversity.
Beta diversity quantifies how different the microbial communities are across various samples.
-
What is alpha diversity and how is it measured? (4)
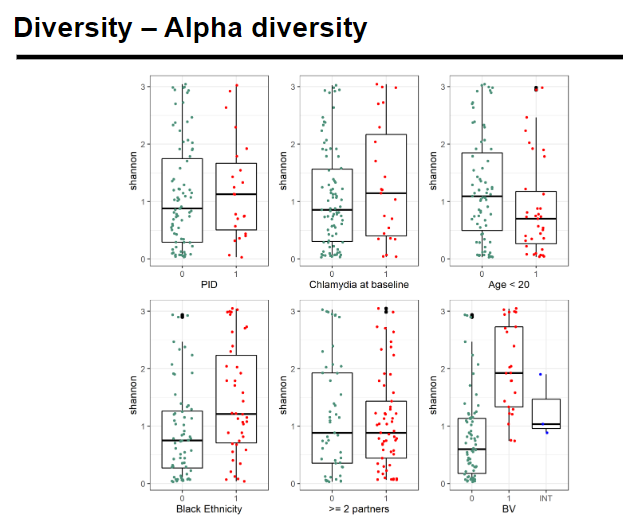
Alpha diversity refers to the diversity within a single sample.
It is often measured using:
Species richness: The number of different species detected in a microbial ecosystem.
OTU count: The number of Operational Taxonomic Units (OTUs) present.
Species diversity: Measured by metrics like the Shannon index, considering both the richness and evenness of species.
Species evenness: Measures how balanced species are in abundance, i.e., whether some species dominate over others.
-
What is beta diversity and how is it measured? (4)
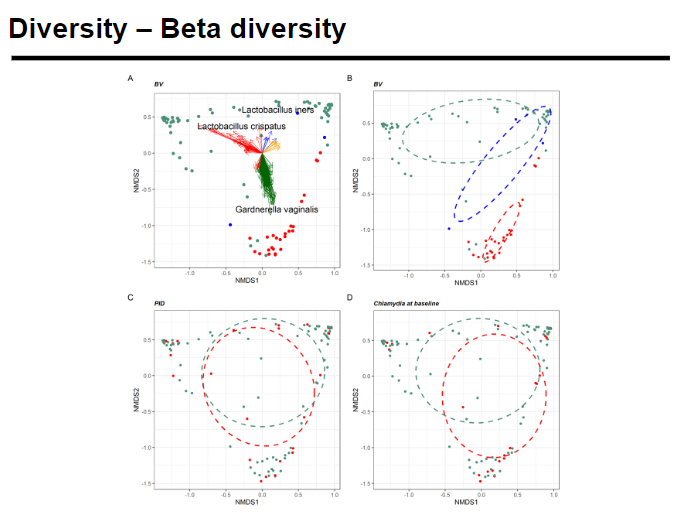
Beta diversity measures how different the microbial composition is between two environments or samples.
It can be quantified using the following metrics:
Bray-Curtis dissimilarity: Based on the abundance of species in the samples.
Jaccard distance: Based on the presence or absence of species, without considering abundance.
UniFrac: Based on the phylogenetic trees of the microbial communities, comparing evolutionary relationships.
-
Picture demonstrating High throughput sequencing:
-
Picture demonstrating High throughput sequencing: - Read length
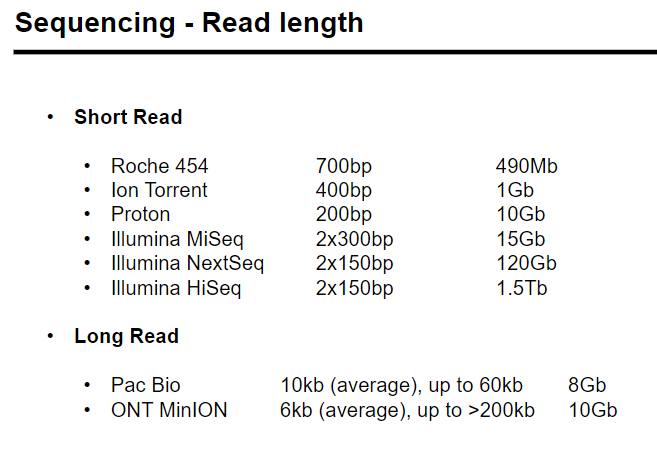
-
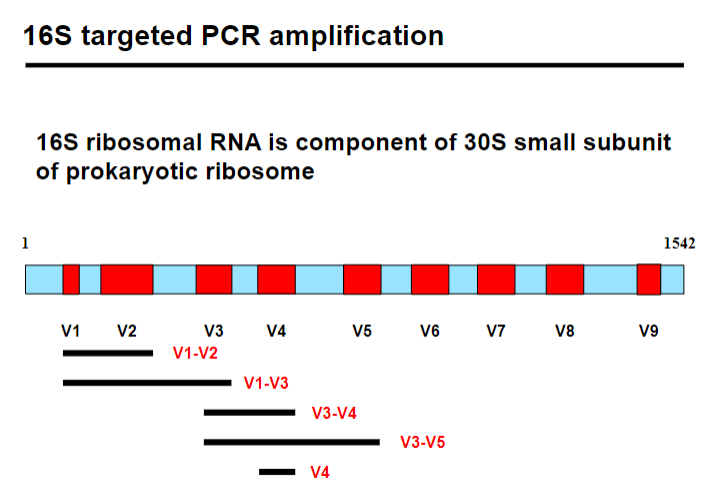
Picture demonstrating 16S targeted PCR amplification:
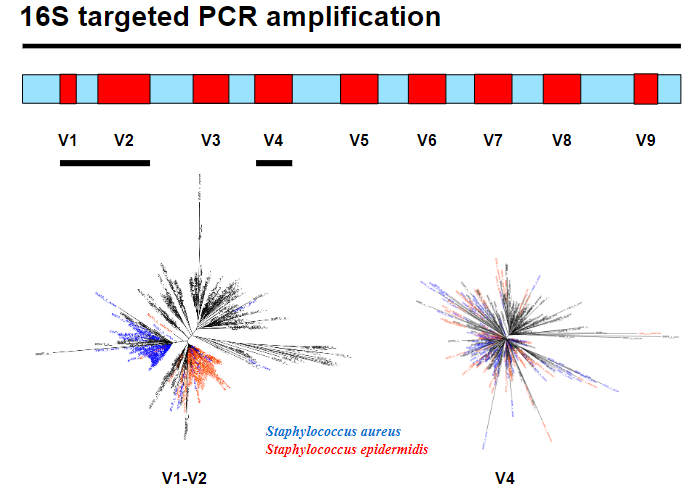
-
Which variable region should be chosen for 16S PCR amplification? (2)
Phylogenetic signal: Choose a region that provides strong evolutionary information, helping distinguish species accurately.
Amplicon length: Consider the length of the amplicon to balance amplification efficiency and sequencing quality.
-
Picture demonstrating 16S targeted PCR amplification:
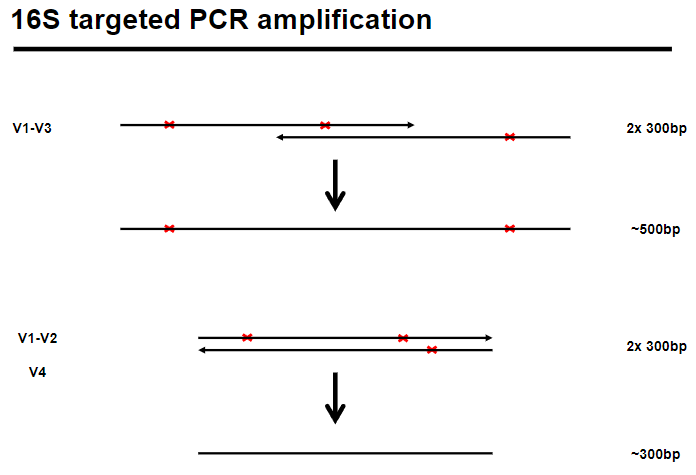
-
What controls should be used in 16S targeted PCR amplification? (4)
16S rRNA gene: Found in all bacteria, making it a universal target for microbial studies.
Contamination risks: PCR is highly sensitive to contamination from various sources, including:
Environment: External contamination from the surroundings.
Operator: Contamination introduced during handling and processing.
Reagents: Contaminants present in reagents used in the PCR process.
Low biomass samples: Contamination risks are especially important for low biomass samples.
Consider using a "kitome" (collection of reagents) control to monitor contamination from commercial kits.
-
Picture demonstrating 16S targeted PCR amplification – kitome:
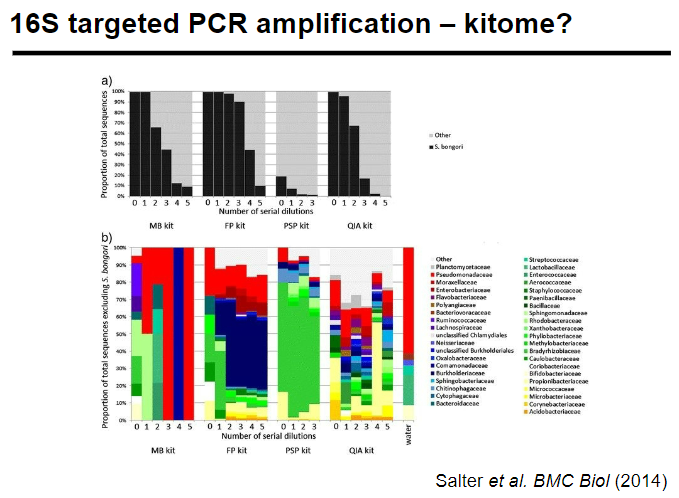
-
How can contamination be mitigated in 16S targeted PCR amplification? (5)
Randomize samples: To reduce systematic bias in sample processing.
Note batch numbers of reagents: Helps track potential contamination from specific reagent lots.
Sequence negative controls: To detect any contamination introduced during PCR.
Mitigate contamination risks: Through proper laboratory practices and handling.
Choice of variable region: Determines the resolution of microbial classification, with less reliability below genus level.
-
What factors affect resolution and reliability in 16S targeted PCR amplification? (4)
Choice of variable region: Determines the resolution of microbial classification, with less reliability below the genus level.
New long-read technologies: Enable full-length 16S sequencing, improving resolution.
PacBio
Nanopore
Error rates: Long-read technologies have higher error rates, introducing noise, although development is ongoing.
-
What are the technological approaches in metagenomics? (2)
Targeted 16S PCR amplification: Used to assess taxonomic diversity in a sample, but biased as it only targets bacteria.
Whole genome shotgun sequencing: Provides a more comprehensive view of microbial diversity, including bacteria, archaea, eukaryotes, and viruses.
-
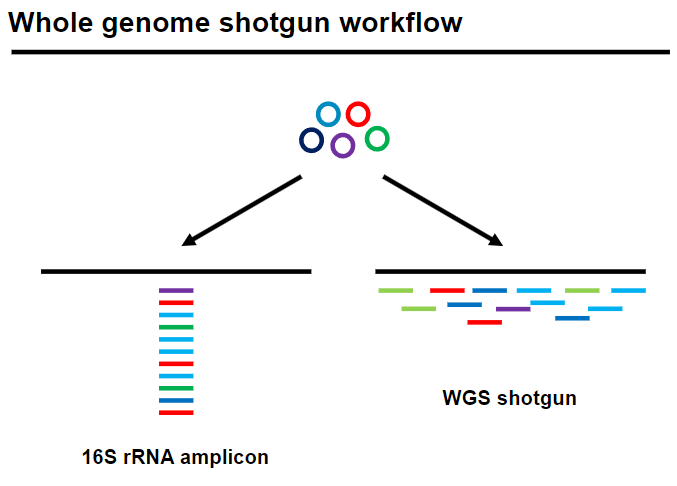
Picture demonstrating Whole genome shotgun workflow:
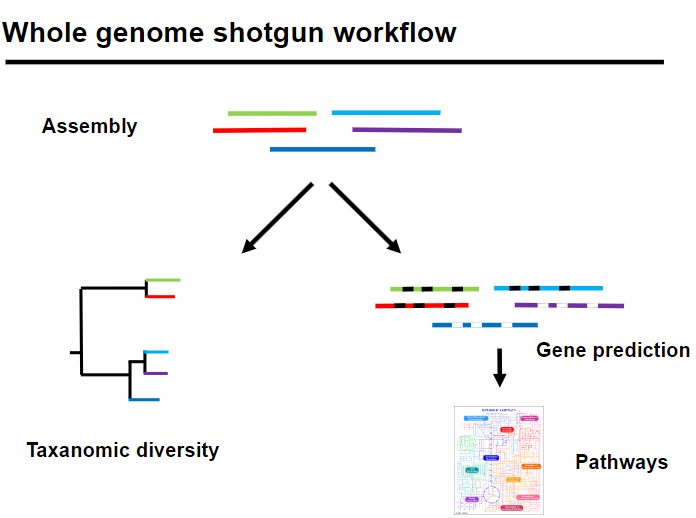
-
What are the challenges in whole genome shotgun sequencing of environmental samples? (3)
Host cells in excess: Host DNA often dominates the sample, complicating the analysis of microbial DNA.
No amplification step: Without an enrichment step, bacterial DNA might be diluted by the host DNA.
Sample-dependent contamination:
Faecal samples: Typically contain <10% human DNA.
Saliva, nasal, skin samples: Can have >90% human DNA contamination.
-
How can enrichment be achieved in whole genome shotgun sequencing without amplification? (3)
Pre-extraction: Use differential lysis to break down mammalian cells, enriching microbial DNA but may bias towards gram-positive bacteria.
Post-extraction:
Enzymatic degradation of methylated nucleotides targets mammalian DNA but can introduce bias against AT-rich bacterial genomes.
-
What are the technological approaches in metagenomics? (3)
Targeted 16S PCR amplification: Assesses taxonomic diversity in a sample but is biased as it targets only bacteria.
Whole genome shotgun sequencing:
Provides an unbiased view of taxonomic diversity.
Assesses composite gene functions in the sample.
Includes all microorganisms, not just bacteria.
-
What is metagenomics and its key benefits? (3)
Metagenomics is the study of genetic material recovered directly from environmental or biological systems/compartments.
It provides an unbiased view of taxonomic diversity in a sample.
Key advantages:
Not limited by the ability to culture organisms.
Offers an overall view of gene content in the sample.

