The Epigenome (Genomics)
SDL: The Epigenome The aim of this session is to introduce the epigenome. This is the complete set of chemical signals in the DNA, and the histones that the DNA is wrapped around, that affect gene expression during development. These epigenetic signals distinguish one tissue from another and can be used to distinguish different types of cancer. They are also thought to be one of the main ways that the environment acts on the genome to affect gene expression. Learning Outcomes On successful completion of the lecture, students should be able to: Discuss the DNA packaging problem and how chromatin is the basis of the solution Describe what the epigenome is Describe the specific epigenetic mechanisms of DNA methylation, histone modification, X-inactivation and genomic imprinting Describe pharmacoepigenetics with respect to epigenetic regulation of genes and the epigenetic effects of drugs Describe the use of epigenetic enzymes as drug targets, using cancer as an example
-
What is the epigenome and why is it important? (3)
The epigenome refers to all the heritable changes in the genome that do not involve alterations in the DNA sequence but affect gene expression.
It is crucial for regulating gene expression.
Epigenetic mechanisms such as DNA methylation, histone modification, X-inactivation, and genomic imprinting control gene expression patterns.
-
How is the genome packaged inside the cell? (3)
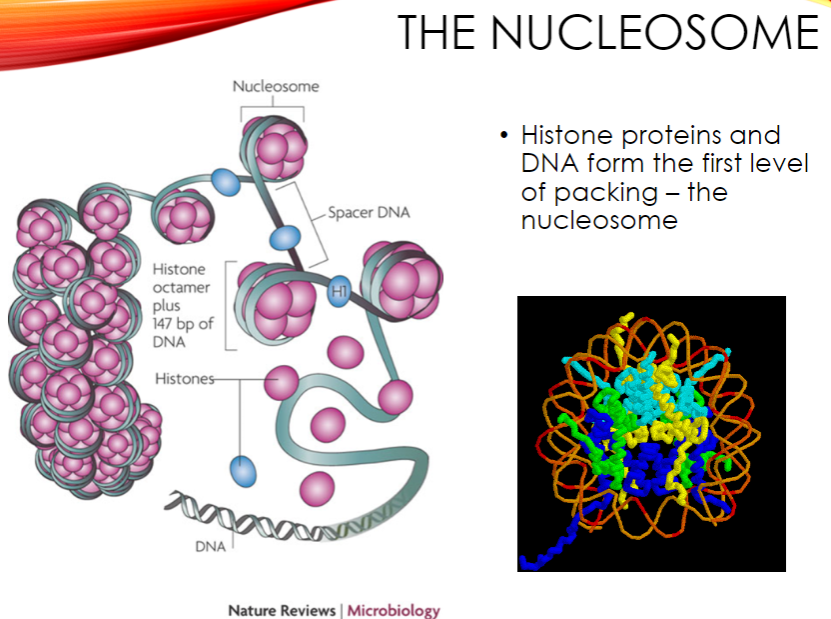
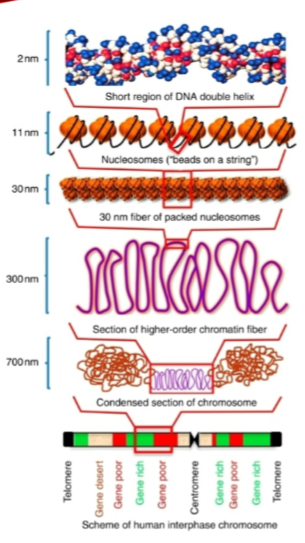
The genome is packaged into the nucleosome, where histone proteins and DNA form the first level of packing.
Nucleosomes are further wound into 30nm fibers.
These fibers are packaged with scaffold proteins to form higher-order structures, with chromosomes being the densest form of genomic DNA.
-
What are the key epigenetic mechanisms? (5)
DNA methylation, which regulates gene expression.
Histone modification, which affects chromatin structure and gene expression.
X-inactivation, which silences one of the X chromosomes in females.
Genomic imprinting, where the expression of certain genes depends on the parent of origin.
Pharmacoepigenetics, which studies how epigenetic changes influence drug response.
-
What is euchromatin and how is it characterized? (3)

Euchromatin is gene-rich and transcriptionally active.
It has a dispersed appearance under a microscope.
It contains unique DNA sequences.
-
What is heterochromatin and how is it characterized? (3)
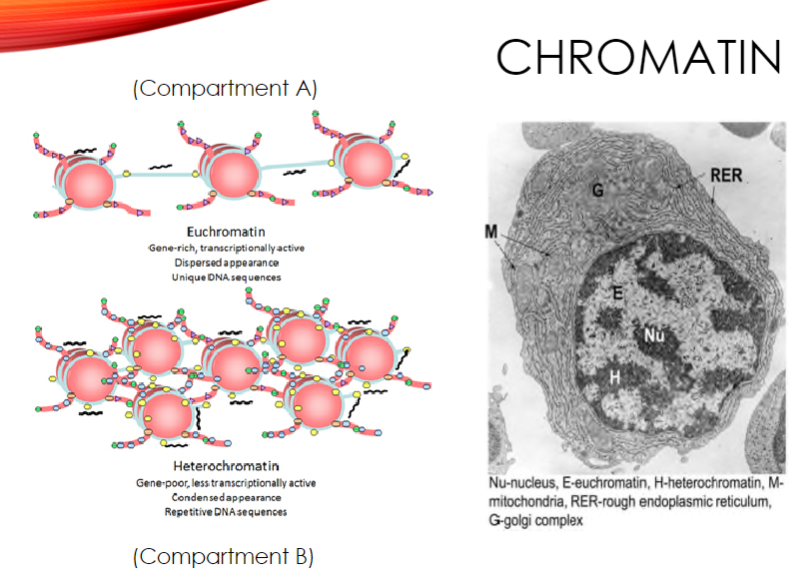
Heterochromatin is gene-poor and less transcriptionally active.
It has a condensed appearance.
It contains repetitive DNA sequences.
-
What is the epigenome? (2)
The epigenome refers to the sum of all heritable changes in the genome that do not occur in the primary DNA sequence.
These changes affect gene expression and result in a change in phenotype but not genotype.
-
What are the key mechanisms of epigenetic regulation? (4)
DNA methylation
Histone modification
X-inactivation
Genomic imprinting
-
What is the nucleosome and its role in chromatin structure? (2)
The nucleosome is the first level of DNA packing, formed by histone proteins and DNA.
It plays a crucial role in the compacting of the genome into higher-order chromatin structures.
-
How are nucleosomes organized in the chromatin structure? (3)
Nucleosomes are wound up to form 30nm fibers.
These fibers are then further wound up with scaffold proteins.
This process generates higher-order structures that lead to the formation of chromosomes.
-
What are the two types of chromatin and their characteristics? (4)
Euchromatin: Gene-rich, transcriptionally active, dispersed appearance, and unique DNA sequences.
Heterochromatin: Gene-poor, less transcriptionally active, condensed appearance, and repetitive DNA sequences.
-
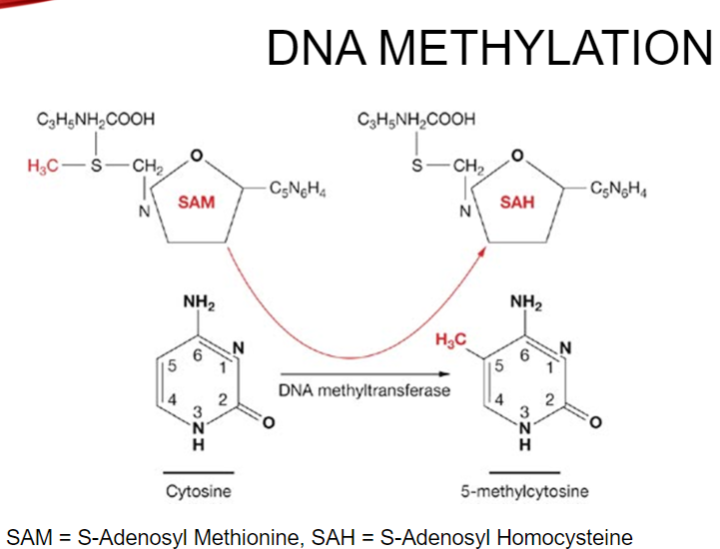
What is DNA methylation and its process in humans? (4)
DNA methylation is the addition of a methyl group to the 5' position of a Cytosine.
This process is catalyzed by DNA methyltransferase enzymes (DNMT1, DNMT3a, DNMT3b).
S-Adenosyl Methionine provides the methyl group required for this modification.
In differentiated cells, DNA methylation occurs in CpG dinucleotides.
-
Picture demonstrating DNA methylation 1:
-
Picture demonstrating DNA methylation 2:
-
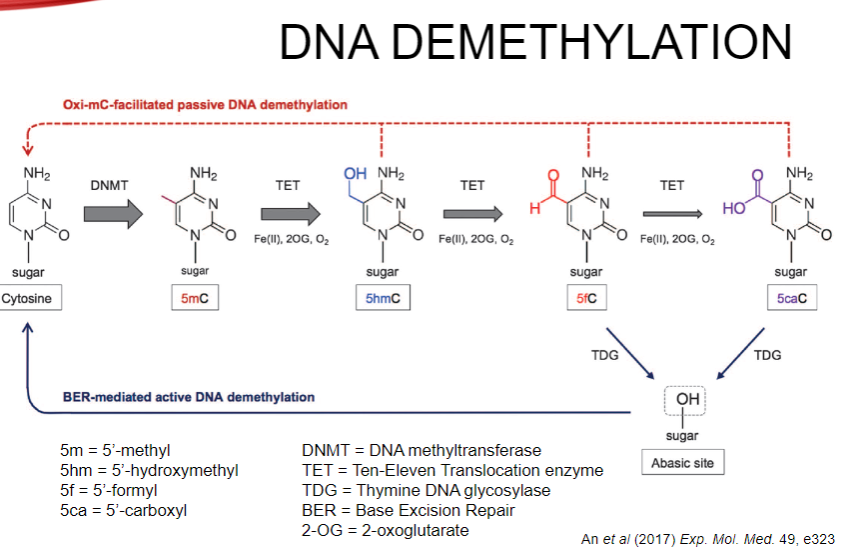
How do DNA methylation patterns change over time? (1)

DNA methylation patterns change during development and play a crucial role in controlling gene expression.
-
What is histone modification? (1)
Histone modification is the addition of chemical groups to the proteins that make up the nucleosome.
-
How many known histone modifications exist and how many of them are well understood? (2)
There are over 100 known histone modifications, and many are of unknown function.
-
What types of enzymes catalyze histone modifications? (1)
A large range of enzymes catalyze histone modifications.
-
What are the common types of histone modifications? (4)
Methylation
Acetylation
Phosphorylation
Ubiquitination
-
What is the significance of histone modifications involving different amino acids? (1)
Many different amino acids can be modified, and they may have 1 to 4 groups added, leading to a large number of modifications.
-
How are histone modifications named? (1)
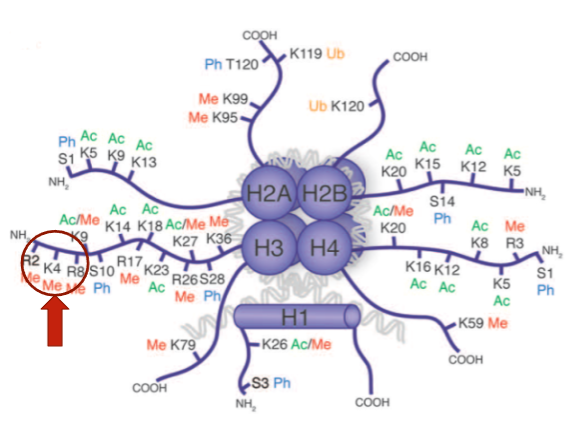
How are histone modifications named? (1)
-
What does the histone modification H3K4Me3 mean? (2)
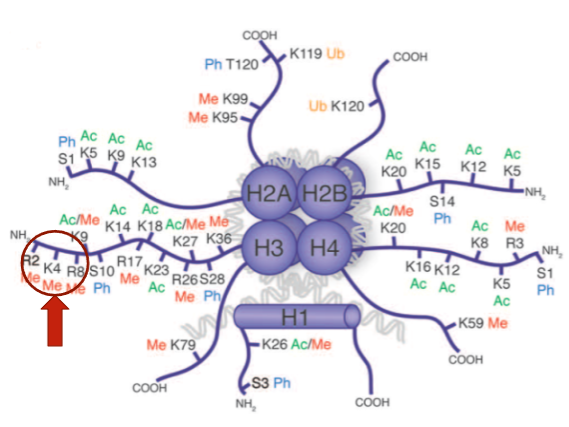
H3K4Me3 means that on Histone 3, the Lysine (K) at position 4 is tri-methylated.
-
What are the categories of histone modifiers? (3)
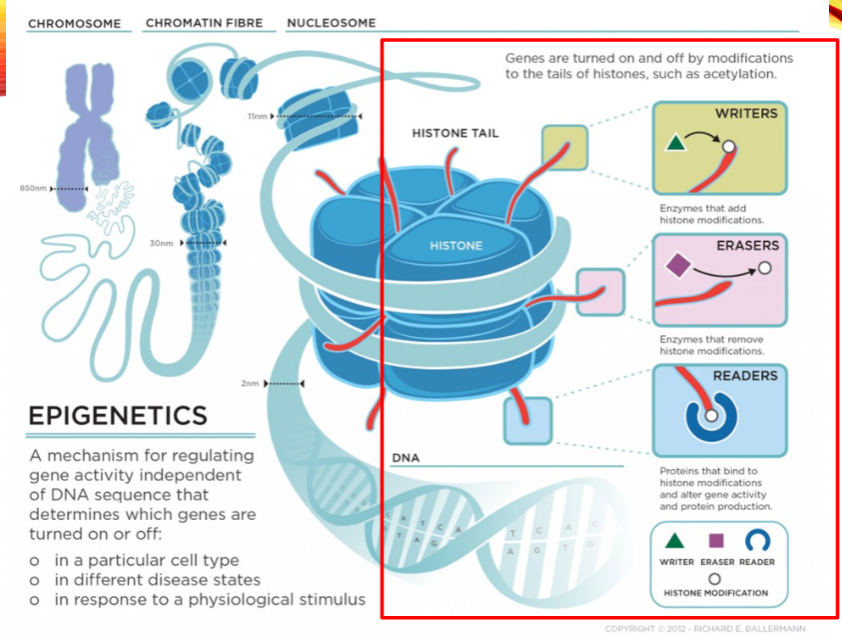
Writers
Erasers
Readers
-
Name two examples of histone writers. (2)
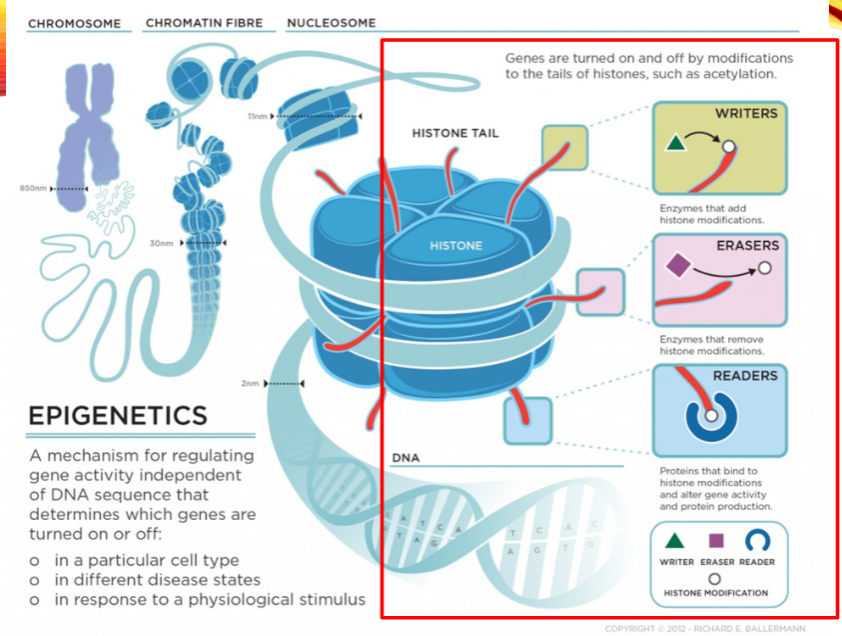
Histone Acetyltransferase (HAT1)
Histone Methyltransferase (EHMT1)
-
Name two examples of histone erasers. (2)
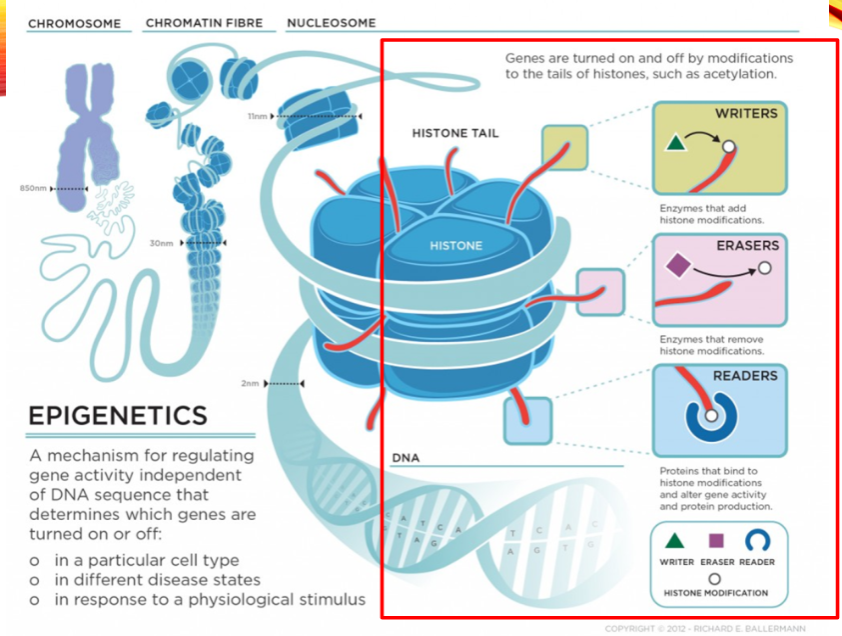
Histone Deacetylase (HDAC1)
Histone Demethylase (KDM1)
-
Name two examples of histone readers. (2)
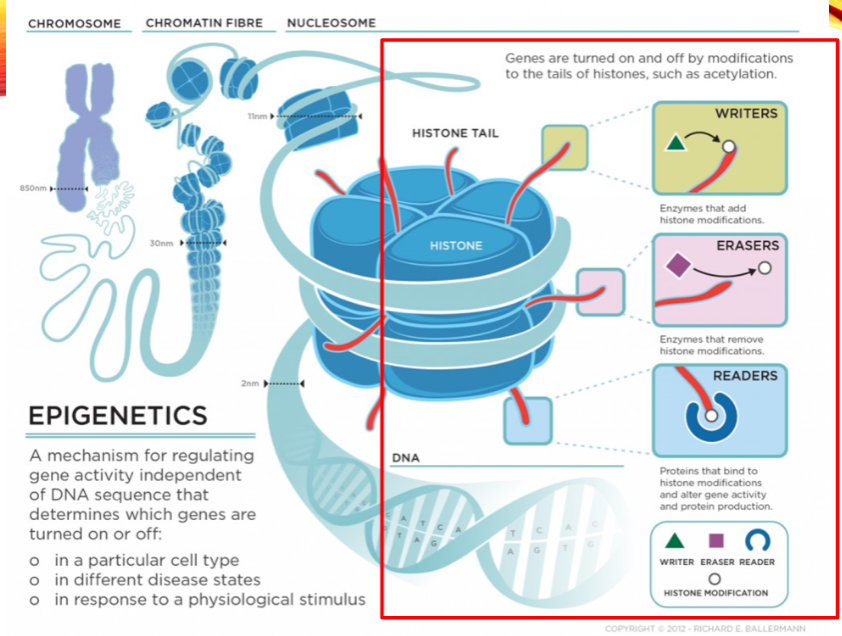
Bromodomain and extra-terminal (BET) proteins (BRD2)
Chromodomain proteins (CBX1)
-
What is the role of histone acetylation? (2)
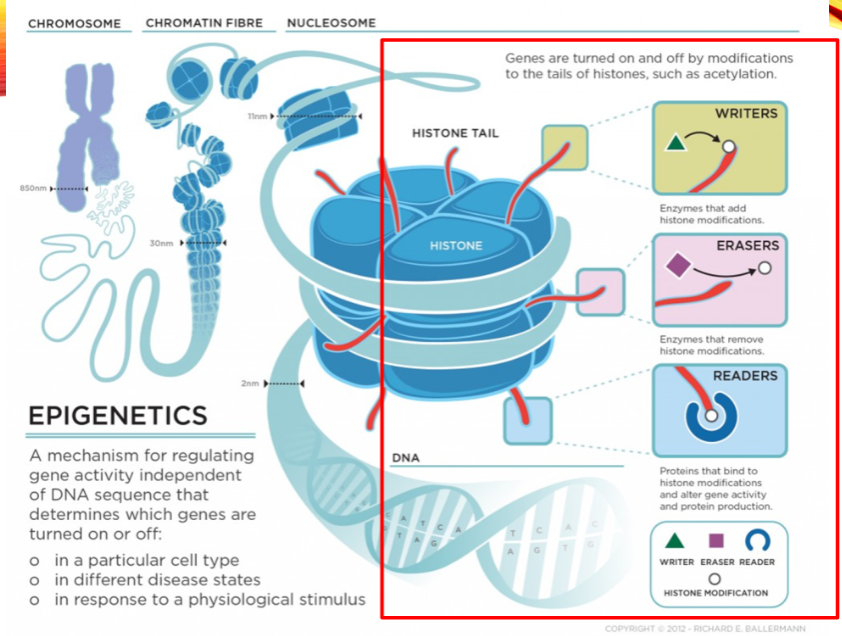
Histone acetylation at Lysine residues relaxes the chromatin structure.
It reduces the positive charge on histones, making it accessible for transcription factors.
-
How does histone methylation affect transcription? (2)
Histone methylation is complex.
It can either repress or activate transcription depending on where it occurs.
-
How do concurrent histone modifications affect gene expression? (1)
Concurrent histone modifications can interact and affect gene expression in complex ways.
-
What is X-inactivation? (1)
X-inactivation is the inactivation of one of the two X chromosomes in every somatic cell in females.
-
Why is X-inactivation necessary? (2)
The Y chromosome has virtually no genes, so there is only one copy of each X chromosome gene in males (hemizygosity).
X-inactivation ensures that every somatic cell in all humans has the same number of active copies of every gene.
-
What is the role of the Xist gene in X-inactivation? (2)
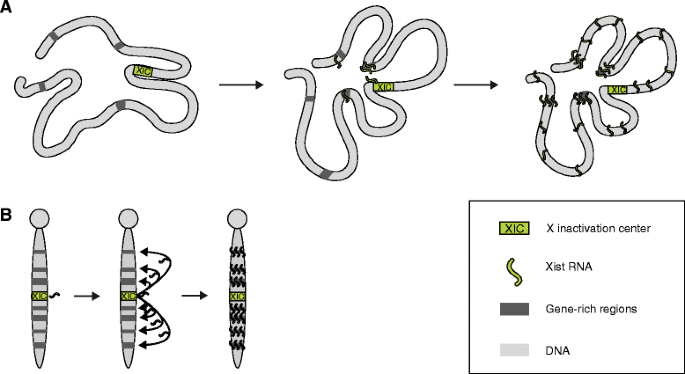
The Xist gene is transcribed as a long noncoding RNA (lncRNA) from the X-inactivation center (Xic).
It binds all over the X-chromosome to initiate X-inactivation.
-
What happens to histones and DNA during X-inactivation? (2)
Histone acetylation is removed.
Histone and DNA methylation occur, contributing to the inactivation of the X-chromosome.
-
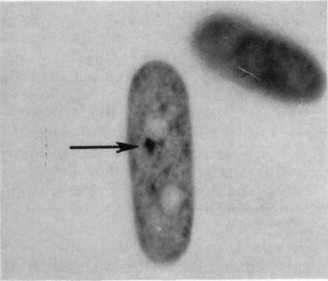
What is the result of X-inactivation at the chromosomal level? (1)
The inactive X-chromosome becomes heterochromatic and forms a Barr body.
-
What is the role of Tsix in X-inactivation? (2)
Tsix is transcribed in the opposite direction to Xist.
It antagonizes Xist RNA to keep one X chromosome active.
-
Why are all tortoiseshell cats female? (1)

Tortoiseshell cats are female because the coat color is determined by alleles on the X chromosome, and females have two X chromosomes.
-
What alleles do tortoiseshell cats have on their X chromosomes? (2)

One X chromosome carries an orange fur allele.
The other X chromosome carries a black fur allele.
-
How does random X-inactivation result in the tortoiseshell pattern in cats? (2)

Random X-inactivation in different cells results in patches of orange fur from one X and black fur from the other X.
This causes the characteristic tortoiseshell coat pattern.
-
What is genomic imprinting? (1)
Genomic imprinting is the selective expression of genes based on the parental origin of the gene copy.
-
What is the basic genetic setup for autosomal genes in humans? (2)
Every autosomal gene has one paternal and one maternal copy.
The expression of some genes is determined by which parent the gene copy is inherited from.
-
Where are imprinted genes typically found? (1)
Imprinted genes are often found in clusters.
-
How many imprinted genes are there in humans? (1)
There are very few imprinted genes, around 250.
-
How is imprinting mediated? (1)
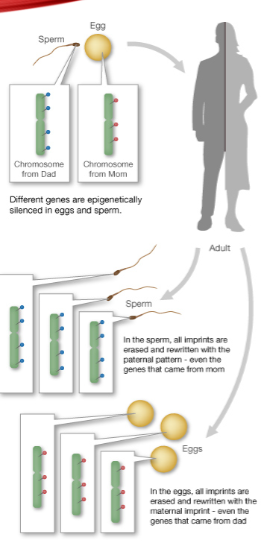
Imprinting is mediated by imprinting control regions (ICRs).
-
How is one copy of an imprinted gene silenced? (2)
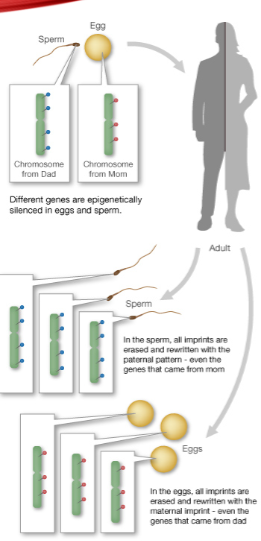
One copy is silenced by DNA methylation, catalysed by DNMT3a.
Histone methylation also contributes to gene silencing.
-
What is the role of LncRNAs in genomic imprinting? (1)
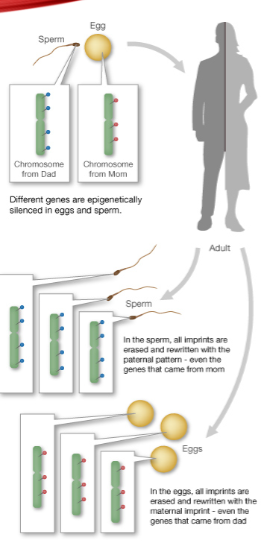
Long noncoding RNAs (LncRNAs) are essential to the imprinting process.
-
When are imprinting patterns reset? (1)
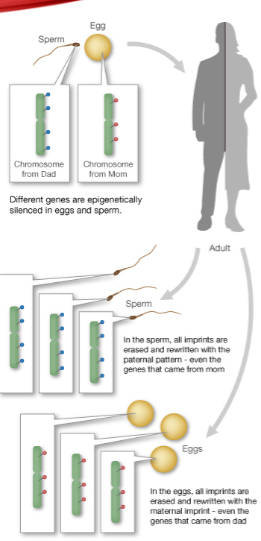
Imprinting patterns are reset during gamete formation.
-
What are the two general mechanisms for epigenetic modification? (2)
DNA methylation.
Histone modification.
-
What are the two specific mechanisms of epigenetic modification? (2)
X-inactivation.
Genomic imprinting.
-
What is the concept of pharmacogenetics and its relevance in cancer treatment? (3)
Pharmacogenetics refers to the study of how genetic variations affect an individual’s response to drugs.
Relevance to cancer treatment: Epigenetic modifications can influence the efficacy of cancer therapies by altering the way genes are expressed in tumour cells.
Big Pharma is interested in targeting epigenetic pathways to improve drug response and potentially develop better cancer treatments.
-
What is pharmacogenetics, and how does it relate to epigenetics in cancer therapy? (4)
Pharmacogenetics focuses on how genetic factors influence drug absorption, distribution, metabolism, and excretion (ADME).
Epigenetics influences gene expression through modifications like DNA methylation, histone modification, etc., without changing the DNA sequence.
In cancer, epigenetic alterations can render tumours more or less responsive to certain treatments.
By understanding pharmacogenetics and epigenetics, more effective drugs can be developed, improving cancer treatment.
-
Can cancer be treated by targeting epigenetic enzymes? (3)
Yes, targeting epigenetic enzymes may offer a new approach in cancer treatment.
Epigenetic enzymes regulate DNA and histone modifications, which are often altered in cancer cells.
Epigenetic therapies aim to reverse these changes, restoring normal gene expression and potentially suppressing tumour growth.
-
What are the key epigenetic changes observed in cancer cells? (4)
Global DNA methylation is altered in tumour cells, impacting gene expression.
Hypermethylation of tumour suppressor genes leads to their silencing, promoting cancer.
Hypomethylation of tumour-activating genes can lead to their overexpression, aiding tumour growth.
Epigenetic enzymes like DNMT3A and TET1/2, which regulate DNA methylation, are often mutated in cancer cells.
-
What are histone modifications and their role in cancer epigenetics? (5)
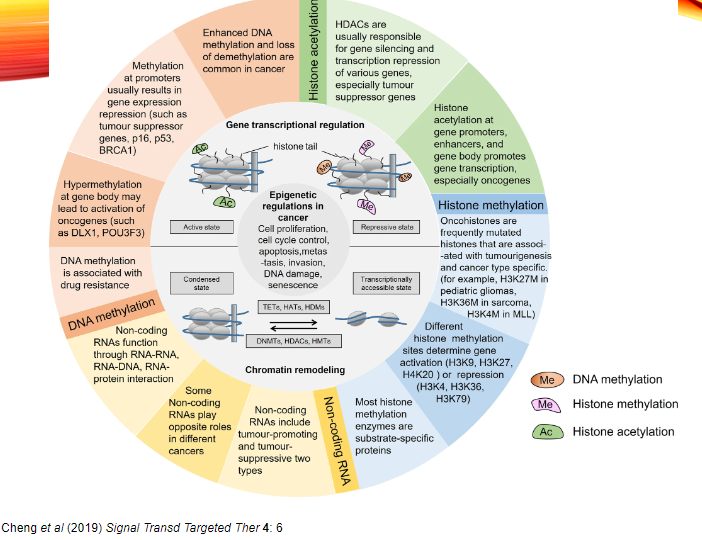
Histone modifications include acetylation, methylation, phosphorylation, and ubiquitination of histone proteins, which regulate gene expression.
Histone Acetyltransferases (HATs) add acetyl groups, often leading to gene activation.
Histone Methyltransferases (HMTs) add methyl groups, which can either silence or activate genes, depending on the specific modification.
Histone Kinases add phosphate groups to histones, affecting chromatin structure and gene expression.
Histone Demethylases remove methyl groups, often activating silenced genes.
-
What is the role of 'histone readers' in epigenetic regulation in cancer? (2)
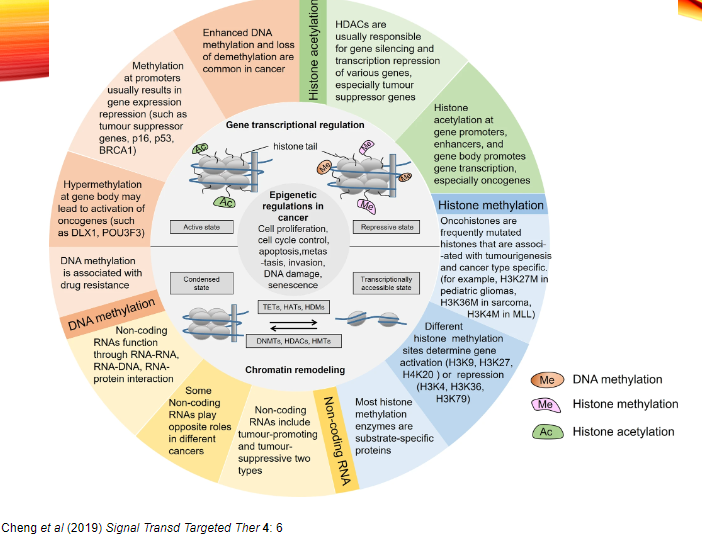
Histone Readers are proteins that recognize and bind to specific histone modifications (e.g., acetyl, methyl, or phosphoryl groups).
In cancer, histone readers can help in interpreting and amplifying epigenetic signals, thus contributing to uncontrolled tumour cell growth.
-
What are some epigenetic enzymes often mutated in tumour cells? (4)
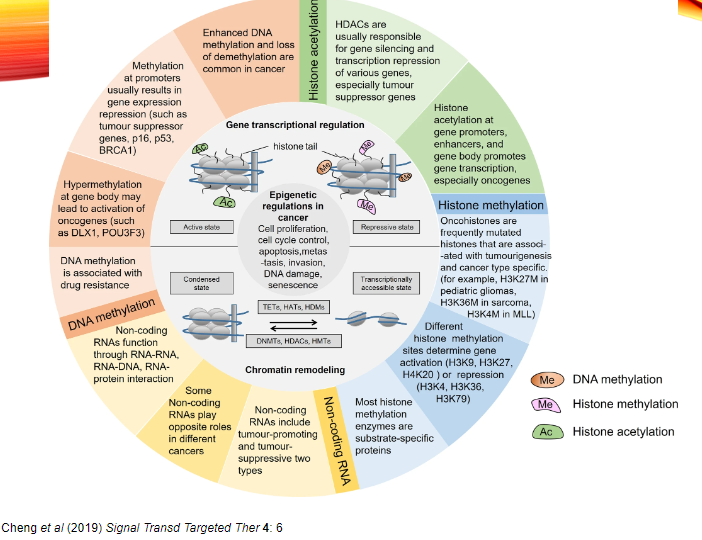
DNMT3A (DNA methyltransferase)
TET1/2 (Ten-Eleven Translocation methylcytosine dioxygenases)
Histone Acetyltransferases (HATs)
Histone Methyltransferases (HMTs)
-
What are DNA Methyltransferase Inhibitors and their role in cancer treatment? (3)
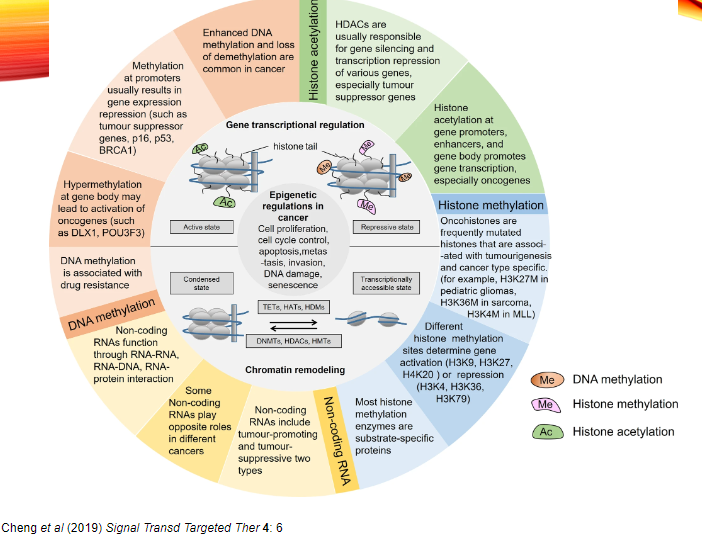
DNA Methyltransferase Inhibitors (DNMT inhibitors) block the activity of enzymes that add methyl groups to DNA, which can silence genes.
Role in cancer treatment: By inhibiting DNMTs, these drugs aim to reactivate silenced tumour suppressor genes, potentially suppressing cancer cell growth.
Example: 5-Azacytidine (Vidaza) is used in the treatment of myelodysplastic syndrome (MDS), a type of cancer affecting blood cells.
-
What is 5-Azacytidine (Vidaza) and its use in cancer treatment? (3)
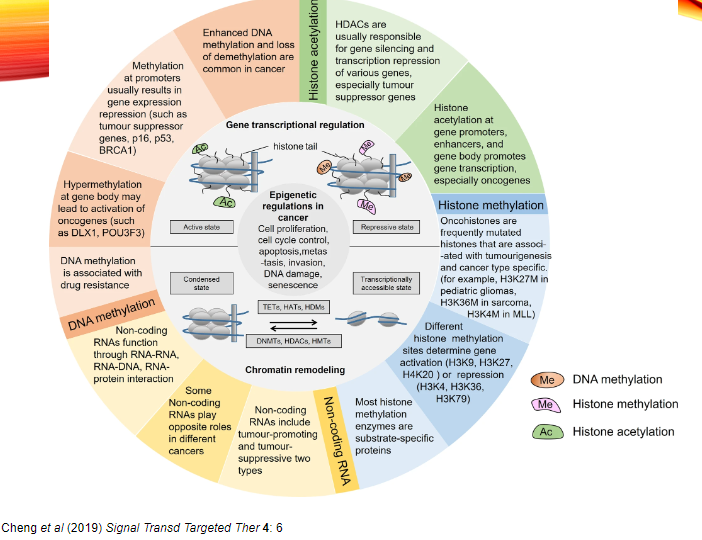
5-Azacytidine (Vidaza) is a DNMT inhibitor that works by blocking DNA methylation.
It is used primarily to treat myelodysplastic syndrome (MDS), a condition that often progresses to leukaemia.
By reactivating silenced genes, it can help prevent the growth of abnormal cells in the bone marrow.
-
What are Histone Deacetylase Inhibitors (HDAC inhibitors) and their role in cancer treatment? (3)

Histone Deacetylase Inhibitors (HDAC inhibitors) block the enzymes that remove acetyl groups from histones, leading to a more relaxed chromatin structure.
This allows tumour suppressor genes to be reactivated, promoting normal cell function and potentially inhibiting tumour growth.
HDAC inhibitors can treat cancers like cutaneous T-cell lymphoma, where gene silencing plays a role in the disease progression.
-
What is Romidepsin (Istodax) and its use in cancer treatment? (3)
Romidepsin (Istodax) is a Histone Deacetylase Inhibitor (HDAC inhibitor) that reactivates tumour suppressor genes.
It is approved for the treatment of cutaneous T-cell lymphoma (CTCL), a type of cancer affecting T-cells in the skin.
By inhibiting HDACs, Romidepsin restores gene expression and can slow the progression of cancer.
-
How many FDA-approved pharmacogenetic drugs are there so far, and what are their uses? (2)
Seven drugs have been FDA-approved so far that target epigenetic mechanisms.
These drugs include DNA Methyl Transferase Inhibitors (e.g., 5-Azacytidine) and Histone Deacetylase Inhibitors (e.g., Romidepsin), and they are used to treat various cancers, such as myelodysplastic syndrome and cutaneous T-cell lymphoma.

