RNA Synthesis (The Living Cell)
At the end of the session, and appropriate background readings, students should be able to: Describe the structure of a eukaryotic gene Describe how RNA transcription is initiated and the role of the general transcription factors Describe different types of RNA polymerase in a eukaryotic cell Describe and understand the purpose of the key RNA processing steps; RNA capping, polyadenylation and splicing Describe alternative splicing and understand its importance Describe how mRNA is exported from the cell nucleus
-
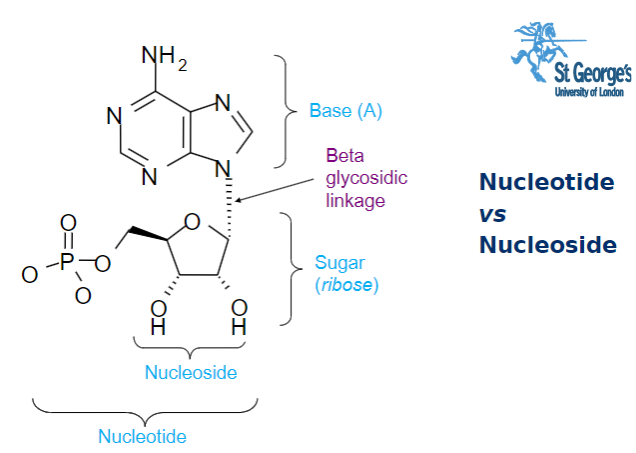
Picture outlining the parts of a nucleotide:
-
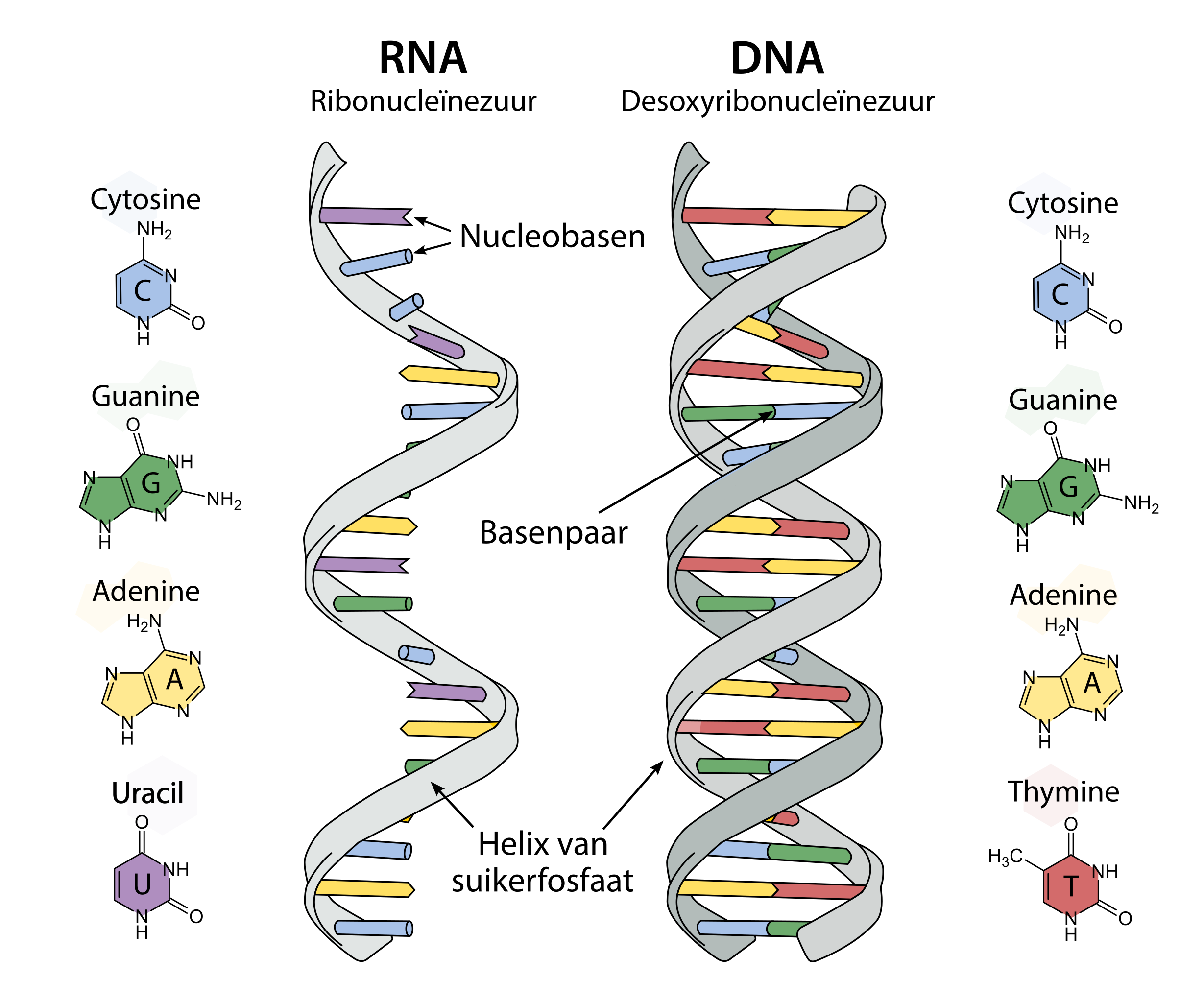
State some basic info about RNA
-Single stranded in eukaryotes. Double-stranded in some viruses where it carries the genetic code
-DNA to RNA to Proteins
-RNA can base pair with DNA, but not capable of forming the B form of the double helix (more A form-like)
-Because RNA single stranded – forms complicated 3D structures
-
What are the 3 forms of DNA? Describe them
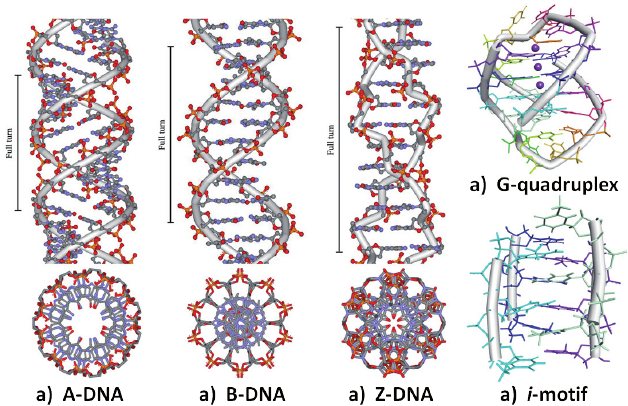
B-DNA:
Most common and biologically relevant form of DNA found in living organisms.
It is a right-handed helical structure with a well-defined and regular geometry.
The B-form is the typical conformation of DNA under normal cellular conditions.
A-DNA:
A-DNA is a more compact and slightly wider helical structure compared to B-DNA.
It can be induced under certain conditions, such as dehydration or in the presence of specific ions.
Z-DNA:
Z-DNA has a left-handed helical structure,
It is characterized by a zigzag or "Z" shape.
-
What are the three major types of RNA? What do they do?
1)Messenger RNA (mRNA)
primary transcript read from DNA
2)Transfer RNA (tRNA)
translation - carries amino acids to ribosome/mRNA complex, the anti-codon on tRNA base pairs with codon on mRNA
3)Ribosomal RNA (rRNA)
Integral to protein synthesis, providing the structural framework for ribosomes where mRNA is translated into proteins.
-
What is coding RNA?
RNA that codes for proteins
-
How much % of RNA is made of coding RNA
In human cells, protein-coding mRNA typically makes up about 1-2% of the total cellular RNA.
In a cell, the majority of RNA is non-coding RNA, which includes ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), and various types of regulatory RNAs such as Small cytoplasmic (scRNA)
-
What are the different types of non-coding RNA and what do they do? (4)
• Ribosomal RNA (rRNA) provides the structural framework for ribosomes where mRNA is translated into proteins
•Transfer RNA (tRNA) transports amino acids to the ribosome for protein synthesis
• Small nuclear RNA (snRNA) found in eukaryotes, is part of the splicing apparatus
• Small nucleolar RNA (snoRNA) involved in methylation of rRNA
• Small cytoplasmic RNA (scRNA) plays a role in the expression of specific genes
-
Homologous chromosomes are made of the two homologs...?
Maternal and Paternal
-
What is a gene?
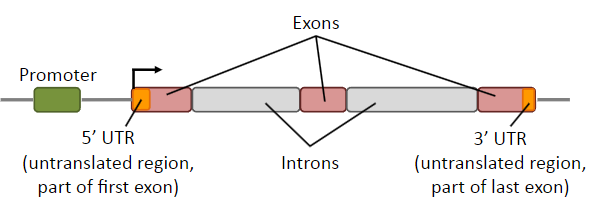
A gene is a DNA segment containing instructions for making aparticular product, including the regulatory elements.
-
What are the two definitions of a gene?
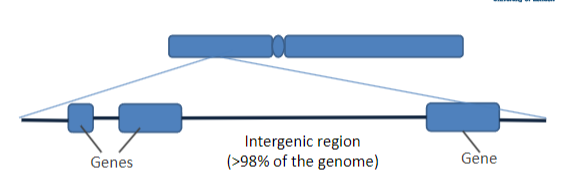
1. Unit of heredity; contains instructions for an organism’s phenotype
2. DNA segment containing instructions for making a particular product
A gene is NOT just the bits that code for a protein
-
Picture demonstrating how genes differ in size:
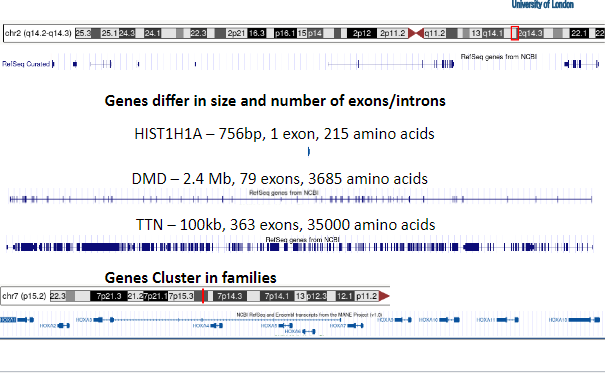
-
What is Transcription? (1 line answer)
Synthesis of mRNA transcript from DNA
-
What is Translation? (1 line answer)
Protein production from mRNA transcript
-
DNA is Transcribed into RNA by RNA Polymerase, what are the 6 steps?
1. Unwinds small portion of DNA
2. Phosphodiester bonds
3. Watson-Crick base pairs
4. Uridine-triphosphate
5. 5’→ 3’ synthesis
6. Single stranded RNA formation
-
What are the 3 main types of Human RNA Polymerases and what genes do they transcribe?
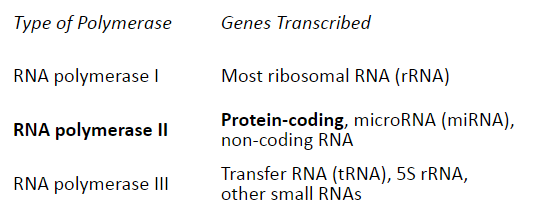
-
What is the 'equipment' needed for RNA synthesis?
DNA template
RNA polymerase II
Ribonucleotides
Buffer
-
What are transcription factors? And where do they assemble?
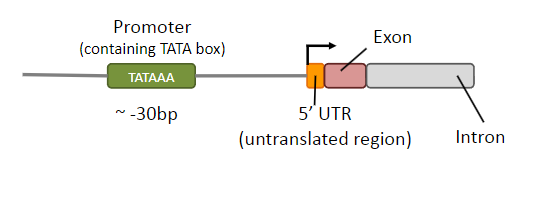
Proteins required to initiate or regulate transcription in eukaryotes
Assemble on promoter to position RNA polymerase II
-
Outline the steps 1-5 on how transcription occurs via assembly of the TATA box (Basal promoter region)
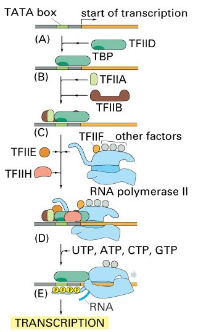
A. TATA box recognised by TATA-binding protein(TBP) subunit of TFIID.
B. TFIIA & TFIIB bind; TFIIA stabilises the complex.
C. Other general transcription factors(E, F, & H) bind, then RNA polymerase II assembles at the promoter, forming the transcription preinitiation complex (PIC).
D. TFIIH pulls apart the DNA helix and phosphorylates RNA Pol II.
E. Phosphorylated RNA Pol II is released from the complex and begins transcription
-
What are the proximal control elements?
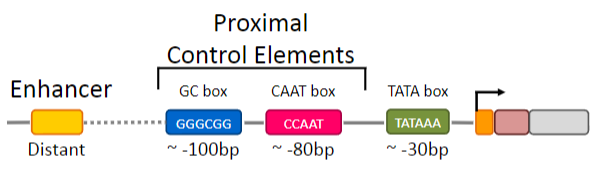
GC Box and the CAAT Box
+Additional upstream sequences are needed for gene-specificregulation of transcription
-
What is the primary transcript?
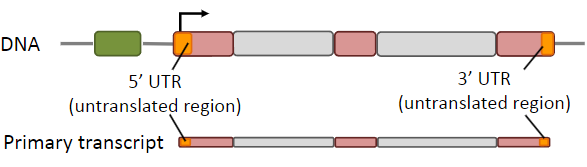
The primary transcript is the initial RNA molecule synthesized from a DNA template during transcription before it undergoes further processing to become a functional mRNA.
-
What are UTRs? And what is the difference between them at the 5' and 3' ends?
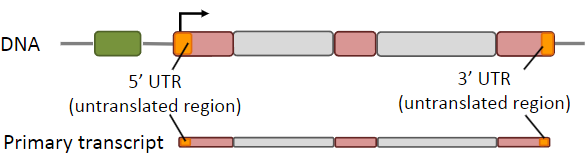
UTRs are transcribed but not translated but play essential roles in the regulation and post-transcriptional processing of mRNA
5’ UTR: regulation of translation
3’ UTR: mRNA stability & miRNA binding
-
What are the 3 main steps of RNA processing?
1)Capping
2)Polyadenylation
3)Splicing
-
What are the 4 steps of capping?
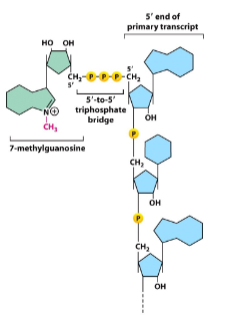
5' end modification:
A modified guanine nucleotide is added to the 5' end of an mRNA, forming a 5'-5' triphosphate bridge.
Guanine nucleotide:
Guanine nucleotide, is methylated at the 7th position.
Capping enzyme complex:
A capping enzyme complex, guanylyltransferase, adds the guanine cap, a methyltransferase, which methylates the guanine.
Co-transcriptional modification:
Capping is co-transcriptional
-
Outline the process of polyadenylation
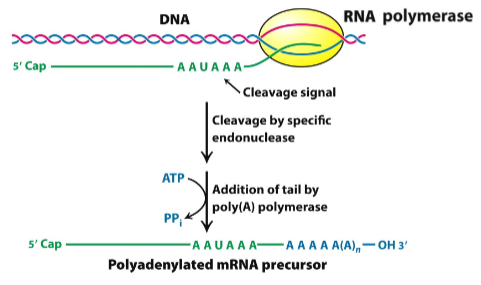
3' end modification:
A string of adenine nucleotides, (a poly(A) tail), is added to the 3' end of an mRNA molecule.
Polyadenylate polymerase:
Poly(A) polymerase adds adenine nucleotides to the mRNA transcript.
Poly(A) tail function:
Enhancing mRNA stability, facilitating nuclear export, and influencing translation efficiency.
Co-transcriptional modification:
Polyadenylation is a co-transcriptional modification.
Recognition signal:
Polyadenylation signal marks the site for poly(A) tail addition
-
Why Cap & Polyadenylate? (3)
1. Stability
2. Transport to cytoplasm
3. Integrity prior to translation
-
What are introns?
Non-coding sequences within genes
-
What are intergenic sequences
Non-coding sequences between genes
-
Outline the process of capping
Introns and exons:
Introns are removed from the pre-mRNA, exons are joined together.
Spliceosome:
It is composed of small nuclear ribonucleoproteins (snRNPs) and other proteins.
Splice donor and acceptor sites:
Where splicing occurs, at the boundaries btt introns and exons.
Alternative splicing:
Alternative splicing allows for the generation of multiple protein isoforms from a gene by selectively including or excluding exons.
Co-transcriptional process:
It is co-transcriptional
-
What is exon skipping?
Certain exons are excluded from the final mRNA transcript, leading to the omission (leaving out) of specific protein-coding sequences
-
What is Intron retention?
In this case, an intron is retained in the final mRNA transcript instead of being spliced out. This intron can potentially contain stop codons, affecting the translated protein.
-
What is the Alternative 5' Splice site?
Different 5' splice sites within an exon are used, resulting in variations in the inclusion of specific sequences at the exon's beginning.
-
What is the Alternative 3' Splice site?
Different 3' splice sites within an exon are utilized, leading to variations in the inclusion of specific sequences at the exon's end.
-
What is mutually exclusive exons?
Two or more exons are present at a particular genomic location, but only one is included in the final mRNA, leading to different protein variants.
-
What is Alternative promoter usage?
Different promoters are used to initiate transcription, resulting in the inclusion of different first exons in the mRNA.
-
What is alternative polyadenylation?
Various polyadenylation sites are used, leading to the inclusion of different 3' untranslated regions (UTRs) in the mRNA.

