Recombinant DNA and Cloning Vectors (Genomics)
SDL: Recombinant DNA and cloning vectors The key principle of recombinant DNA technology is that DNA molecules from different sources can be cut or altered and re-joined and thus used to express recombinant proteins in heterologous cells. In this lecture we will explore some of the reasons why this technology is so powerful but will focus on the use of plasmids in a number of different senarios to express recombinant proteins. The application of recombinant technology include the ability to express medically useful proteins such as human insulin, interferons and other biopharmaceuticals in simple prokaryotes such as E.coli or eukaryotic cell culture. Recombinant technology is already being extended to include gene therapy through the use of gene editing whereby a functional gene can be inserted into an individuals DNA substituting defective genes to rescue a phenotype. In addition to these “gain-of-function” applications scientists are increasingly exploring technologies to engineer “loss-of-function” attributes on genomes with the result that our ability to study gene function is much enhanced and rapidly expanding. Learning Outcomes On successful completion of the lecture, students should be able to: Give examples of various recombinant vectors in molecular biology Plasmids Bacteriophages Viruses Artificial Chromosomes Describe in detail the features of plasmids that make them useful for manipulating fragments of DNA and expression of recombinant proteins, with examples Describe the use of inducible promoters in plasmids Describe the relevant differences between prokaryotic and eukaryotic gene expression requirements Describe the use of gene fusions to improve purity of recombinant proteins
-
What are the types of recombinant vectors used in the molecular toolkit? (4)
Plasmids
Bacteriophages
Viruses
Artificial Chromosomes
-
What are some features of plasmids and phages as recombinant vectors? (4)
Plasmids:
Found in many, but not all, bacteria.
Have a restricted host range.
Transferable by transformation and conjugation.
Phages (Lambda):
Bacterial viruses.
Transfer antimicrobial resistance through transduction.
-
What are the uses of viruses as recombinant vectors? (2)
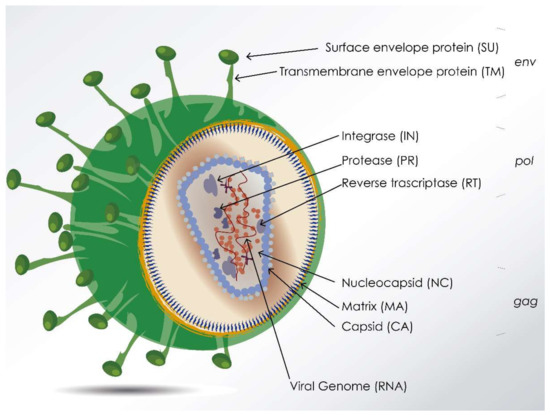
Non-primate Lentiviruses (in picture): Integrate DNA into mammalian cells.
Baculoviruses: Used in combination with recombinant expression in insect cells (eukaryotic system).
-
What are yeast artificial chromosomes (YACs) used for in recombinant DNA technology? (1)
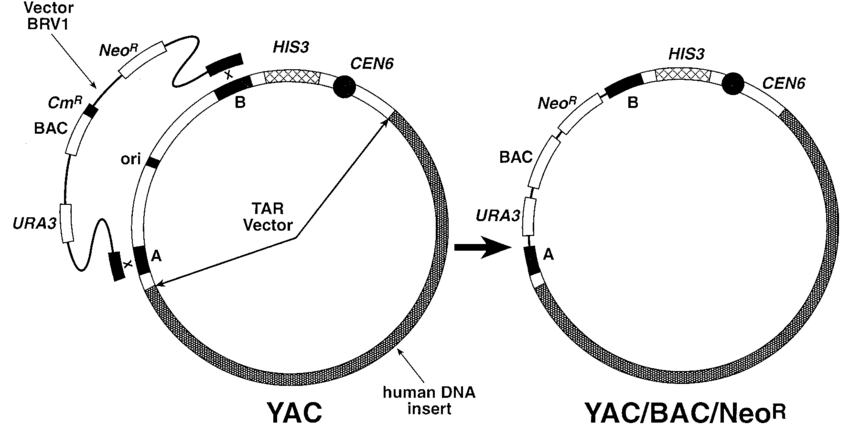
YACs: Used for introducing large segments of DNA.
-
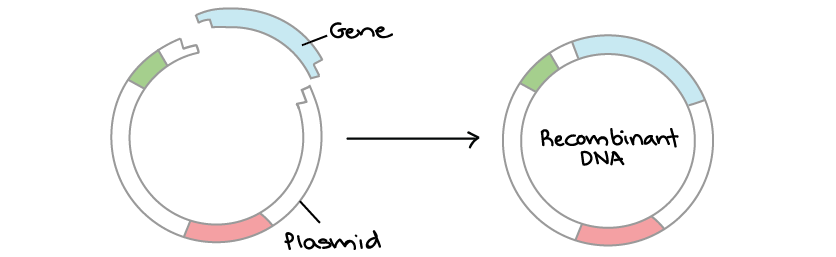
What are plasmids and their significance in bacteria? (4)
-Plasmids are discrete, circular double-stranded DNA molecules found in many, but not all, bacteria.
-They are a means by which genetic information is maintained in bacteria.
-Plasmids exist and replicate independently of the bacterial chromosomes, making them extra-chromosomal.
-They can be exchanged between bacteria, usually within a restricted host range, such as with plasmid-borne antibiotic resistance.
-
What are vectors, and how are they used in molecular biology? (2)
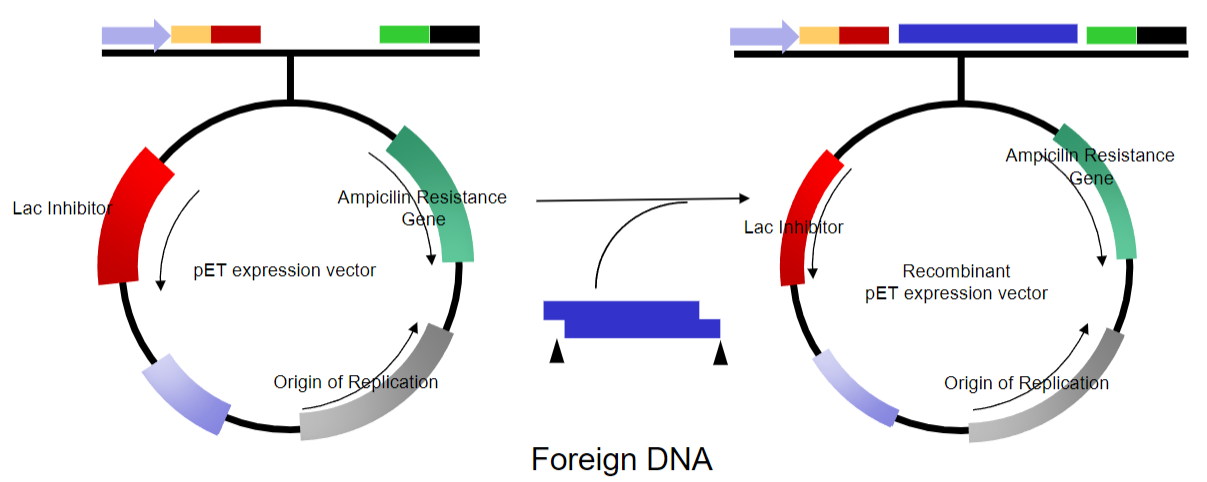
-Vectors are a simplified version of naturally occurring plasmids.
-They are used as molecular tools to manipulate genes.
-
What are the key features of plasmid vectors? (6)
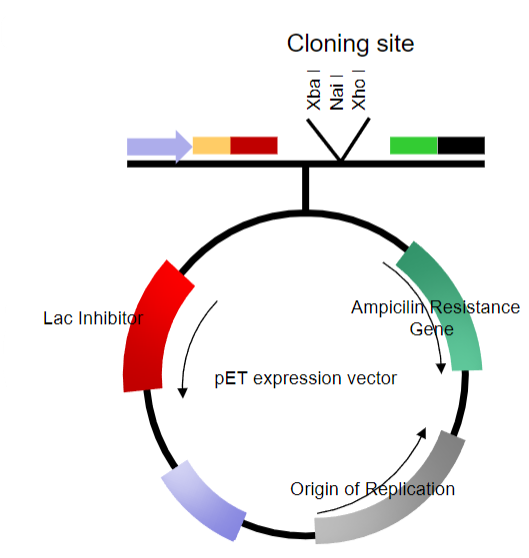
Can be linearized at one or more sites in non-essential stretches of DNA
Can have DNA inserted into them
Can be re-circularized without loss of the ability to replicate
Often modified to replicate at high multiplicity (copy number) within a host cell
Contain selectable markers
Most are relatively small, 4-5kb in size
-
Picture demonstrating Bacterial Plasmids as a vector:
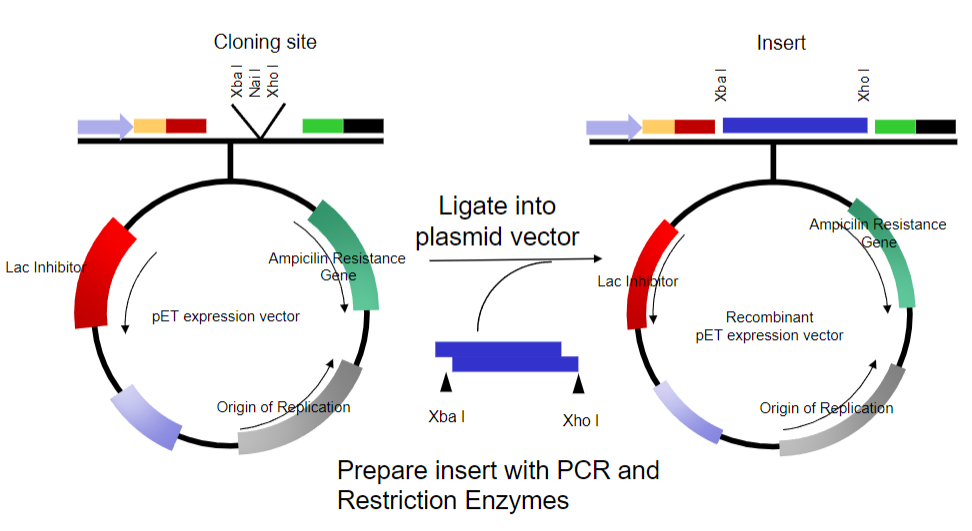
-
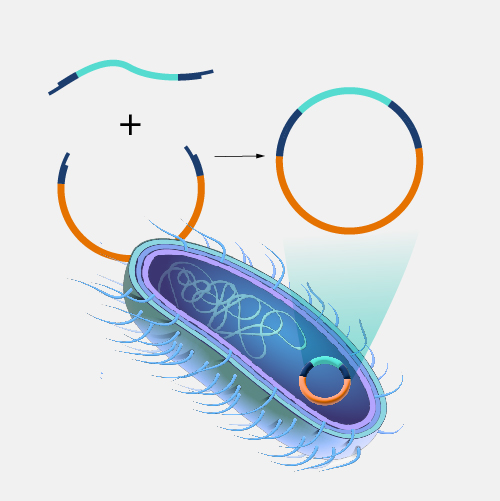
Picture demonstrating Recombinant Proteins fromrecombinant DNA:
-
Why use plasmids as recombinant tools? (2)
Allows for the expression of a recombinant gene in a living organism of choice (prokaryote or eukaryote)
-
How do plasmids add functionality over simple DNA in experimental or functional genomics? (5)
Alter the properties of the gene product
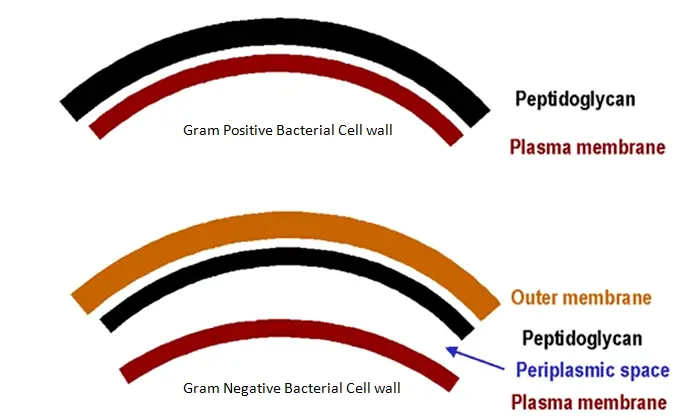
Make it secreted extra-cellularly or into the periplasmic space
Fuse it to a peptide tag or other protein
Make it useful as a therapeutic
Add or modify control elements (e.g., make it inducible or express it to high levels on demand)
-
What do recombinant vectors facilitate the production of? (1)
Recombinant drugs
-
What percentage of all biopharmaceuticals do recombinant proteins or peptides constitute? (1)
About 30%
-
List examples of recombinant proteins used in clinical treatments. (5)
-Human insulin for diabetes
-Interferons-a & b for viral Hepatitis or MS (multiple sclerosis)
-Erythropoietin for kidney disease or anaemia
-Factor XIII for haemophilia
-Tissue plasminogen activator (TPA) for embolism or stroke
-
How many recombinant drugs were approved by the FDA for clinical use between 2011 and 2016? (1)
Around 62
-
When did recombinant antibodies (biologics) first appear in the clinic? (1)
In the late 1980s
-
What percentage of FDA approvals since 2014 were biologics? (1)
30%
-
What percentage of the 62 recombinant drugs approved by the FDA for clinical use between 2011 and 2016 were biologics? (1)
Around 50%
-
How many biologics were approved in 2017, and how does this compare to the previous annual average? (2)
17 biologics were approved in 2017, double the previous annual average.
-
What were the estimated sales of biologics in the US in 2010? (1)
$18.5 billion
-
What percentage of biologics were humanised antibodies in 2021? (1)
57%
-
What is Synagis used for and what type of modification does it have? (2)
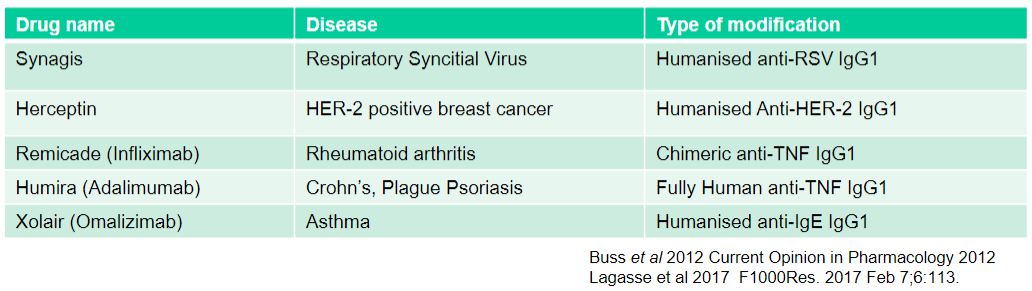
Used for: Respiratory Syncytial Virus
Type of modification: Humanised anti-RSV IgG1
-
What is Herceptin used for and what type of modification does it have? (2)
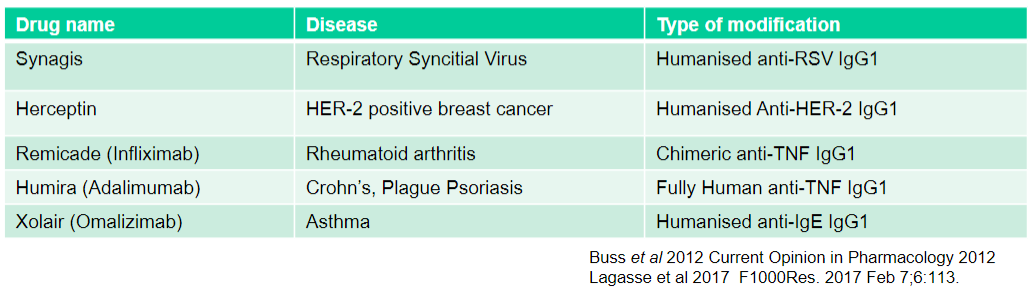
Used for: HER-2 positive breast cancer
Type of modification: Humanised Anti-HER-2 IgG1
-
What is Remicade (Infliximab) used for and what type of modification does it have? (2)
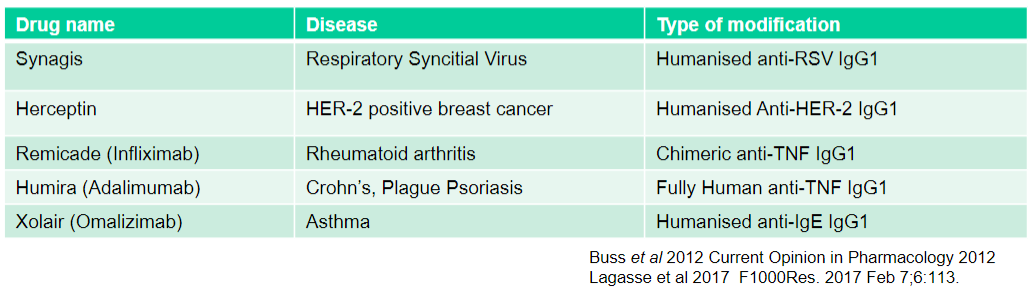
Used for: Rheumatoid arthritis
Type of modification: Chimeric anti-TNF IgG1
-
What is Humira (Adalimumab) used for and what type of modification does it have? (2)
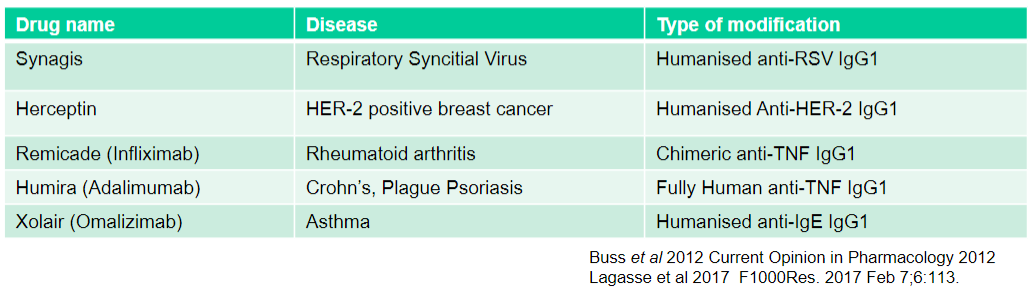
Used for: Crohn's, Plague Psoriasis
Type of modification: Fully Human anti-TNF IgG1
-
What is Xolair (Omalizumab) used for and what type of modification does it have? (2)
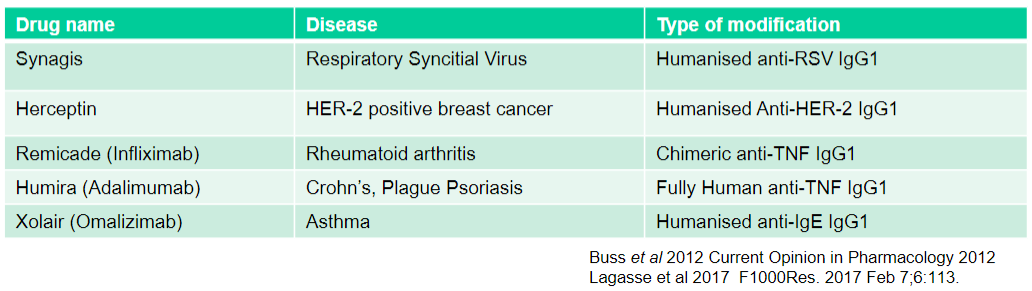
Used for: Asthma
Type of modification: Humanised anti-IgE IgG1
-
What is the objective of cloning the defective gene from a patient with an inherited condition? (1)
To express it in bacteria in large amounts for functional analysis of the protein.
-
What are the requirements for a plasmid in a prokaryotic system to clone the defective gene? (7)
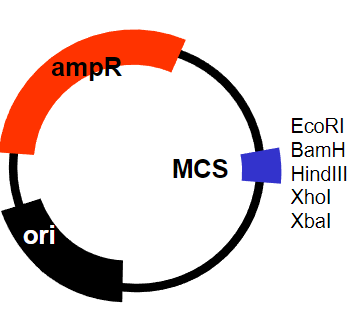
Ability to replicate in bacteria (E. coli)
Maintained at high copy number
Modified origin of replication
Selectable marker containing an antibiotic
Ampicillin resistance gene
Easy to manipulate – cut and re-join
Multiple cloning site (MCS)
-
Why is the gene coding sequence alone insufficient for expression in bacteria? (1)
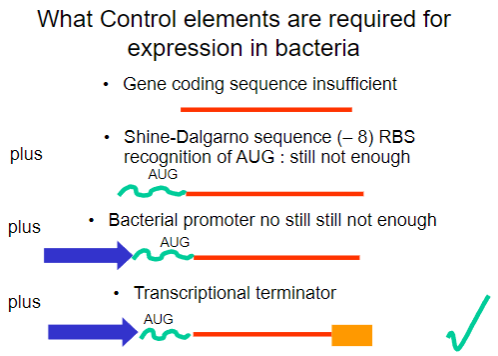
It requires additional control elements for proper expression.
-
What is the Shine-Dalgarno sequence and why is it important? (1)
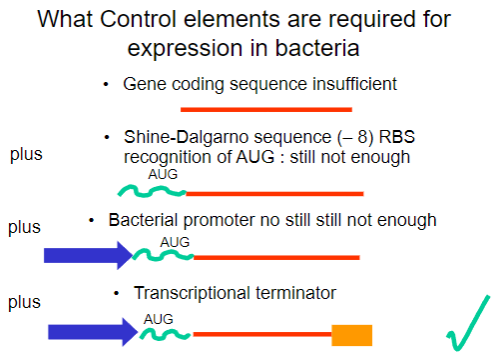
It is a ribosomal binding site (RBS) that facilitates recognition of the AUG start codon.
-
What is required in addition to the Shine-Dalgarno sequence for successful expression in bacteria? (1)
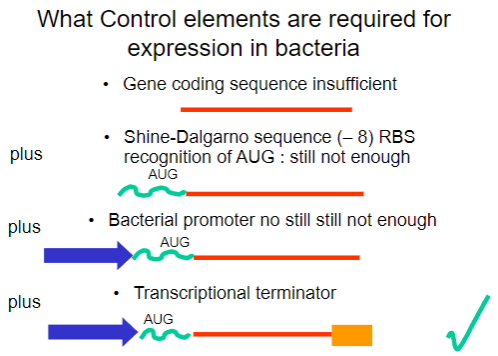
A bacterial promoter is also necessary.
-
What additional control element is required beyond the bacterial promoter for gene expression? (1)
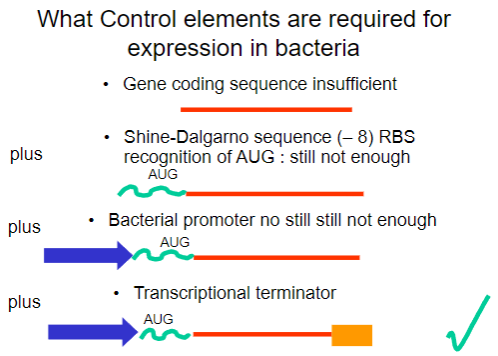
A transcriptional terminator is required to signal the end of transcription.
-
What are the two types of promoters used in bacterial gene expression? (2)
Constitutive - Genes or promoters that are continuously expressed, regardless of environmental conditions.
Inducible - Genes or promoters that are only expressed in response to specific signals or conditions (like the presence of an inducer). They can be turned "on" or "off" as needed.
-
What is a constitutive promoter? (2)
The constitutive promoter always remains on, allowing continuous expression of the foreign gene.
However, if the protein is toxic to the bacteria, the bacteria might stop growing or die, not because the promoter turns off, but because of the detrimental effect of the protein itself on the cells.
-
What is an inducible promoter? (3)
An inducible promoter controls the expression of a gene, and it is only activated in response to an external signal, such as a specific chemical or molecule.
Before activation, the gene under the control of this promoter is not expressed or is expressed at very low levels.
When the appropriate inducer (like a sugar, drug, or other small molecule) is introduced, it binds to a regulatory protein, which then interacts with the promoter to initiate gene expression.
-
What is a disadvantage of using an inducible promoter in cloning vectors? (1)
If the protein expressed by the gene is toxic to the cells, the cells might not survive long enough for the promoter to work as intended.
-
What regulates transcription in inducible promoters like the lac operon? (1)
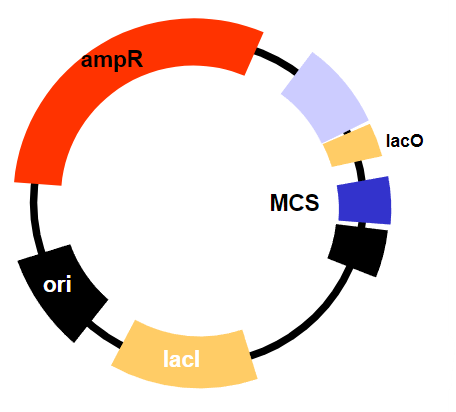
The lac operator, which is blocked by the lac repressor but is de-repressed (released from the repressor) when lactose (or allolactose) is present, allowing transcription to occur.
-
How do inducible promoters utilize transcriptional repressors? (1)
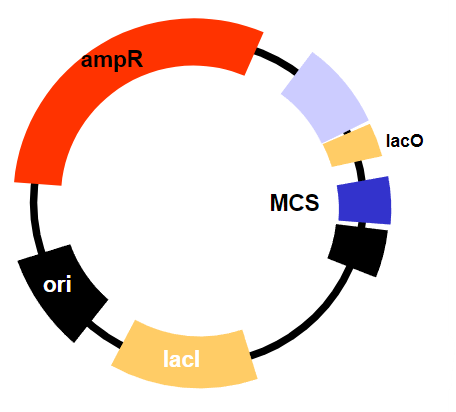
They use transcriptional repressors to control the expression of the target gene.
-
What is required for the function of inducible promoters? (1) and why? (1)
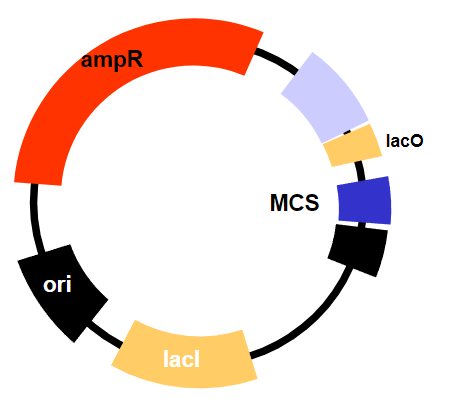
The constitutive expression of the lac repressor (lacI), which normally inhibits transcription but can be removed by the addition of an inducer (like IPTG), allowing transcription to occur.The lac repressor is always present (constitutively expressed) and inhibits transcription until an inducer is added, which causes the repressor to release from the operator and allow transcription to proceed.
-
How is the lac Operator de-repressed experimentally? (1)
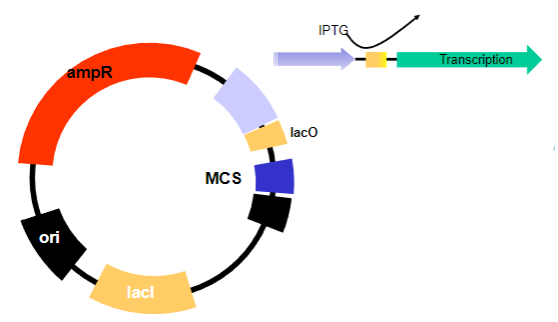
By the addition of a lactose mimic called IPTG.
-
What do inducible promoters typically use to regulate transcription in bacteria? (1)
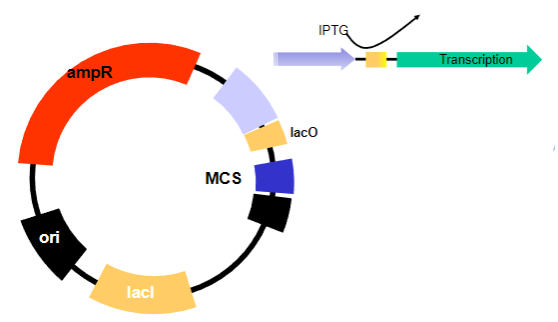
The lac Operator, which is de-repressed by the addition of IPTG.
-
How do inducible promoters utilize transcriptional repressors? (4)
In the absence of the inducer, a transcriptional repressor (often a protein) binds to the inducible promoter or a nearby operator region, blocking the transcription machinery from accessing the gene.
This repression prevents the gene from being transcribed and prevents expression of the protein.
When the inducer (a small molecule or chemical) is introduced, it binds to the repressor. This binding causes a conformational change in the repressor, which dislodges it from the promoter region.
Once the repressor is removed, the transcription machinery can access the promoter and begin transcribing the gene, leading to expression of the target gene.
-
What is required for the function of inducible promoters? (1)
The constitutive expression of a repressor protein (such as the lac repressor) that can be inhibited by an inducer to allow gene expression.
-
What should the DNA insert contain for cloning? (1)
A copy of the coding sequence generated by methods such as PCR.
-
What is required for the DNA insert to facilitate manipulation? (1)
The DNA must be easy to manipulate, allowing for cutting and re-joining with other DNA.
-
What essential elements must the DNA insert include? (2)
The start codon and the stop codon.
-
What type of sequences should the DNA insert consist of? (1)
Only exonic sequences, as bacteria cannot splice introns
-
Is a Cap site required for the DNA insert? (1)
No, a Cap site is not required.
-
Are eukaryotic UTRs required for the DNA insert? (1)
No, eukaryotic UTRs are not required.
-
Is a polyadenylation signal required for the DNA insert in bacteria? (1)
No, bacterial RNAs are not polyadenylated, so a polyadenylation signal is not required.
-
Why are some proteins best made in eukaryotic systems? (2)
Many pharmacologically useful proteins are heavily modified and will not be appropriately processed in bacteria.
Eukaryotic systems allow for post-translational modifications, such as glycosylation.
-
What is an example of a protein that requires eukaryotic expression for proper processing? (1)
Interferons are an example, as they are typically modified by glycosylation.
-
What happens to some proteins when expressed in bacteria? (1)
Some proteins may retain biological activity, while others do not.
-
What is the solution for expressing proteins that require specific modifications? (1)
The solution is to express them in a eukaryotic system.
-
What requirements are already met for the expression in human cells? (6)
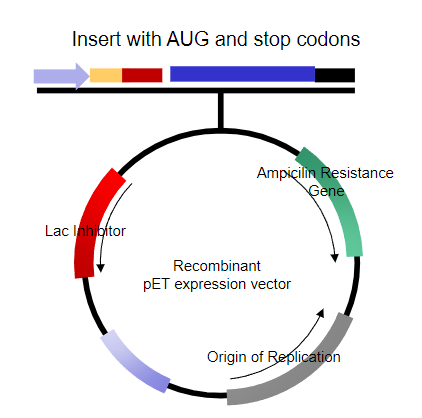
Inducible Promoter.
Shine-Dalgarno sequence.
Insert with in-frame start and stop codons.
Transcriptional terminator.
Origin of replication.
Selectable marker.
Choice of unique restriction sites (MCS).
-
What problems arise when trying to express the protein in human embryonic fibroblasts? (6)
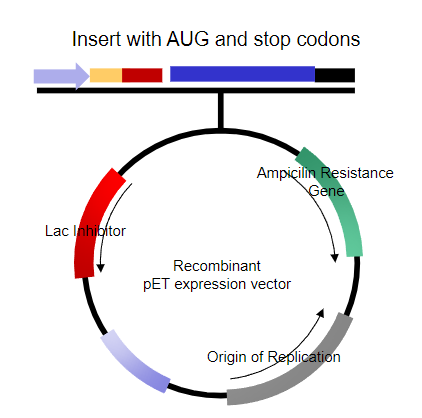
Bacterial promoter doesn’t work in eukaryotic cells.
Shine-Dalgarno sequence isn't recognized in eukaryotic systems.
Transcriptional start is not recognized, and there's no cap site.
No polyadenylation signal for proper mRNA processing.
Termination of transcription is not recognized by eukaryotic RNA polymerase II.
The origin of replication doesn’t function, and it may not be necessary in eukaryotic systems.
-
Why does the bacterial promoter not work in human cells? (1)
Eukaryotic cells require specific eukaryotic promoters for gene expression.
-
What is the significance of the Shine-Dalgarno sequence in this scenario? (1)
It is critical for ribosomal binding in prokaryotes but is not utilized in eukaryotic translation initiation.
-
What is the role of a cap site in mRNA processing? (1)
It is essential for mRNA stability, export from the nucleus, and recognition by ribosomes during translation.
-
Why is polyadenylation important in eukaryotic cells? (1)
It is necessary for mRNA stability and translation efficiency, helping to protect the mRNA from degradation.
-
Picture demonstrating the comparison between prokaryoticand eukaryotic expression vectors:
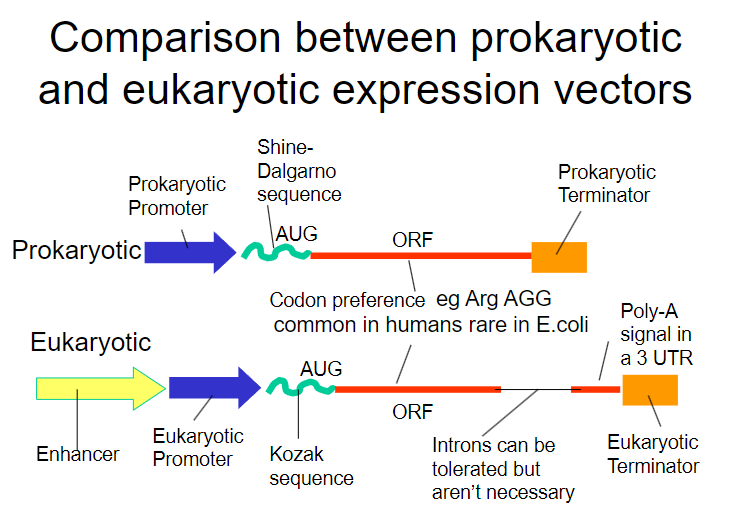
-
What are the requirements for plasmids transfected into a eukaryotic system for transient or stable expression? (4)
The plasmid must be able to replicate within mammalian cells.
Alternatively, the plasmid should integrate into the cell's chromosomes.
The plasmid should contain a selectable marker to identify transfected cells.
The plasmid should be capable of enabling either transient or stable gene expression, such as in the creation of a transgenic cell line.
-
Why is the ability to replicate in mammalian cells important for plasmids? (1)
It allows the plasmid to produce the desired protein efficiently within the host cells.
-
What is the significance of integrating plasmids into chromosomes? (1)
It provides stable, long-term expression of the gene of interest in the host cell.
-
Why is a selectable marker necessary in a eukaryotic system? (1)
It enables the identification and selection of cells that have successfully taken up the plasmid.
-
What is the difference between transient and stable expression in a eukaryotic system? (2)
Transient expression: Short-term expression of the gene, often for experiments requiring temporary protein production.
Stable expression: Long-term expression, achieved by integrating the plasmid into the host genome, resulting in a permanent transgenic cell line.
-
Picture demonstrating Plasmids in a dual systemEukaryotic/Prokaryotic hybrid vector:
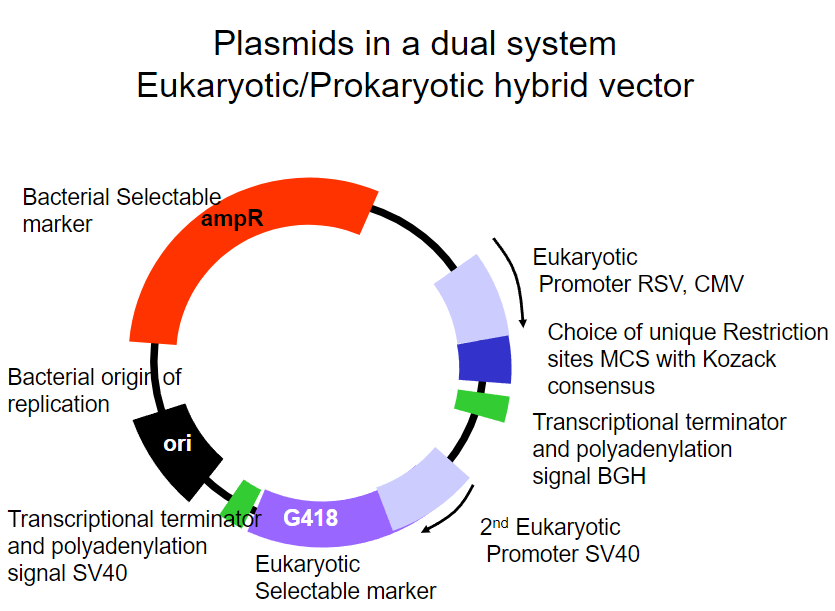
-
What is the challenge faced when expressing proteins in bacteria? (1)
The expressed protein is often difficult to obtain in a pure enough form for functional analysis.
-
What is one potential solution to ease the purification of proteins expressed in bacteria? (1)
Use gene fusions to facilitate purification.
-
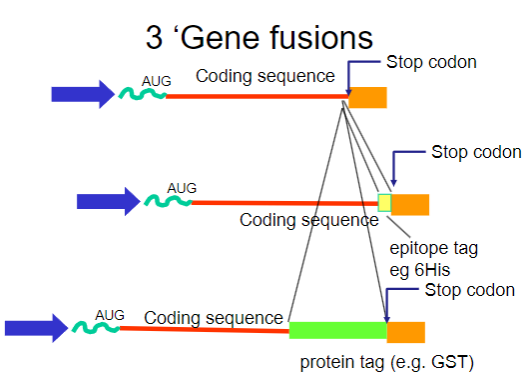
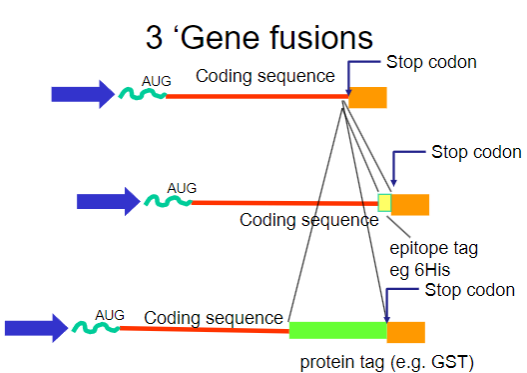
What are gene fusions? (2)
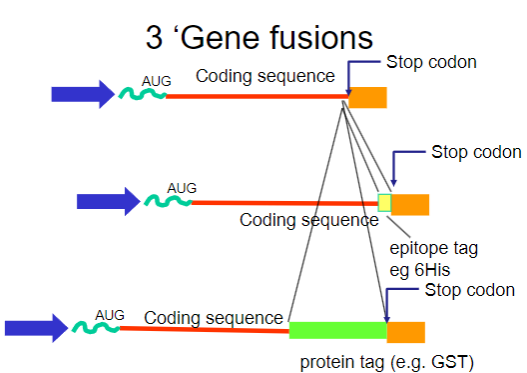
Gene fusions involve combining the coding sequence of the target protein with that of a fusion partner.
The fusion partner often contains a tag that can be easily purified using affinity chromatography.
-
What are common tags used in gene fusions for purification? (2)
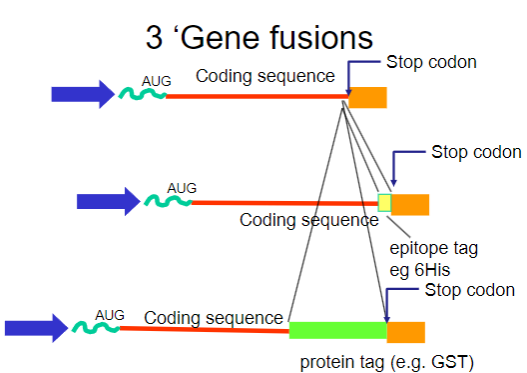
His-tag: A short stretch of histidine residues that can bind to nickel ions in purification columns.
GST-tag: Glutathione S-transferase, which binds to glutathione, allowing for easy purification.
-
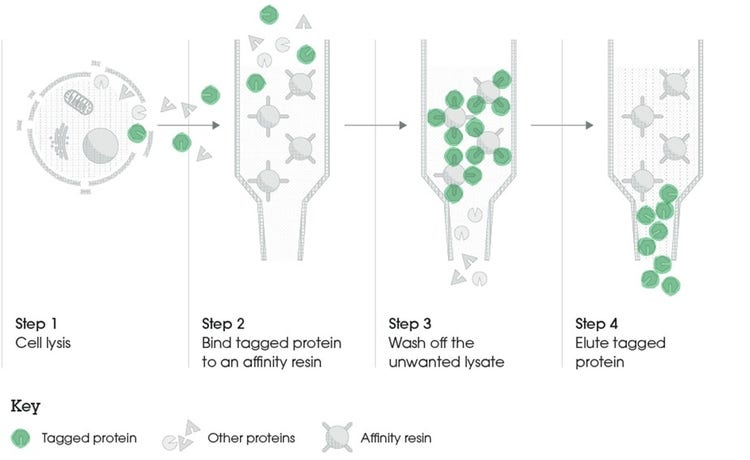
How do gene fusions aid in the purification process? (2)
Gene fusions involve attaching a small, recognizable tag (such as a His-tag) to the protein of interest, which helps the fusion protein bind specifically to an affinity column.
This binding enables the separation of the fusion protein from other proteins in the sample. After purification, the tag can be removed if needed, leaving behind the pure protein for further analysis.
-
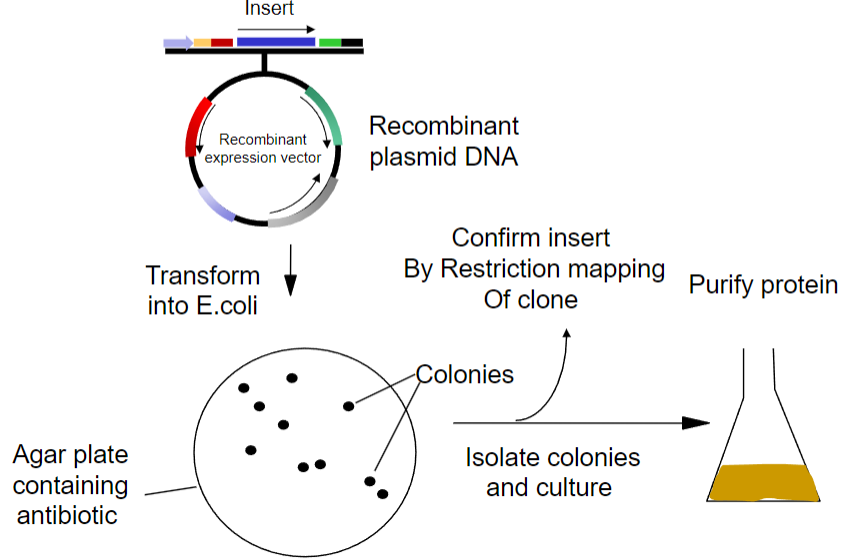
What is the first step in obtaining recombinant proteins from recombinant plasmid DNA? (1)
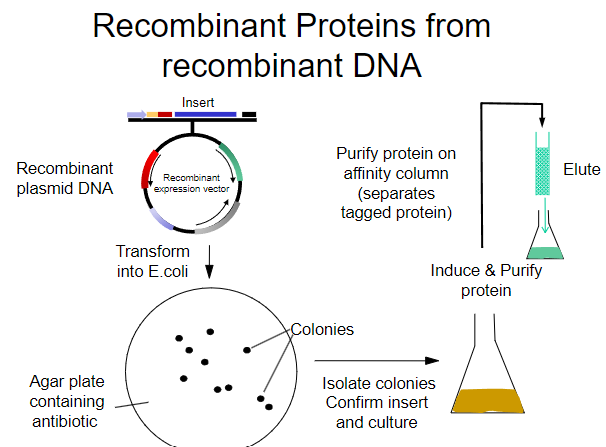
Transform the recombinant plasmid DNA into E. coli.
-
What is the purpose of plating transformed E. coli on an agar plate? (1)
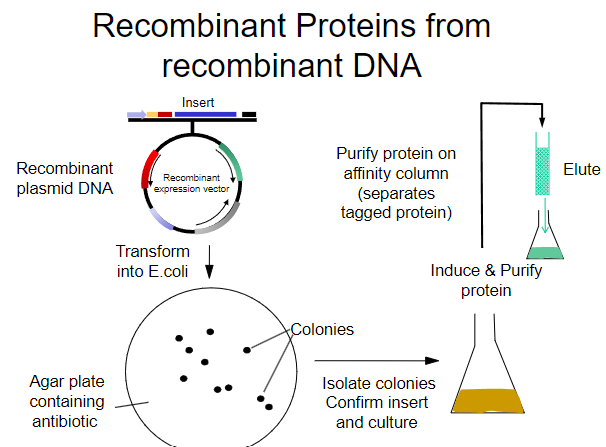
To select for cells that have taken up the plasmid containing the antibiotic resistance gene.
-
What must the agar plate contain for selection? (1)
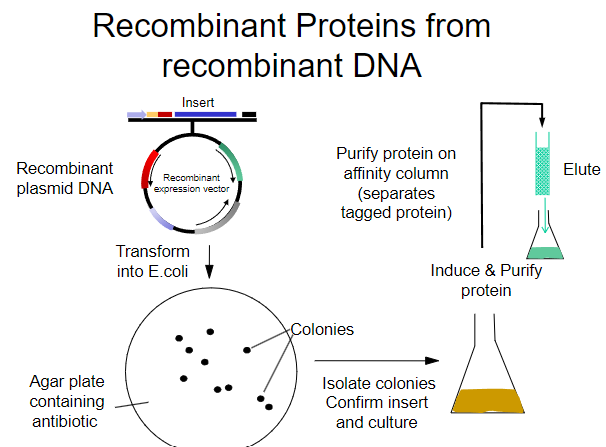
An antibiotic to select for successfully transformed cells.
-
What is the next step after selecting colonies on the agar plate? (1)
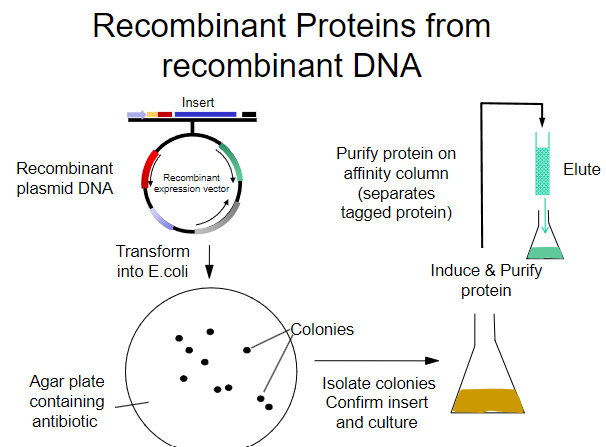
Isolate colonies that contain the recombinant plasmid.
-
What is the process to purify the protein from the isolated colonies? (1)
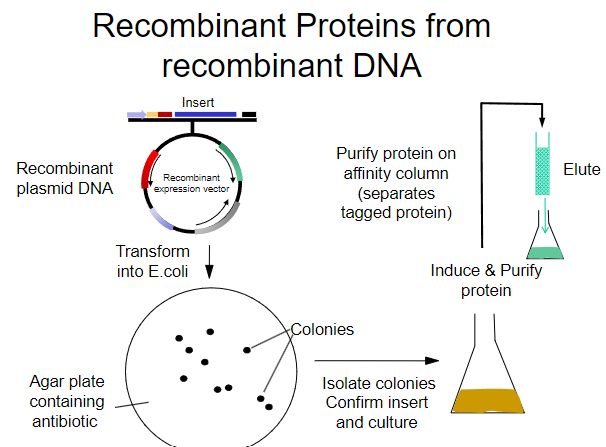
Purify the protein using an affinity column.
-
Which cell type is used for studying the localization of the protein? (1)
Human embryonic fibroblasts.
-
What question does the scenario aim to answer about the protein's location? (1)
Where the protein is located in the cells: cytoplasmic, in the nucleus, or in a membrane.
-

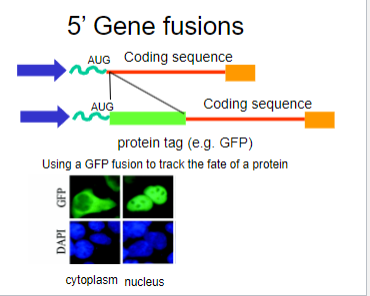
What is Green Fluorescent Protein (GFP)? (1)

A fluorescent protein identified and cloned from jellyfish that emits green fluorescence
-

When was Green Fluorescent Protein (GFP) first identified? (1)

In 1971.
-

What are the properties of GFP that make it useful in biological studies? (2)

It is non-toxic and biochemically inert, allowing for visualization without interfering with cellular processes.
-
Picture demonstrating the use of GFP with gene fusions: