PCR and its Role in Diagnostics (Genomics)
SDL: PCR and its role in diagnostics The theory of DNA melting annealing and hybridisation covered in the previous lecture is also crucial to the concept, design and application of the polymerase chain reaction (PCR) that is at the heart of many modern diagnostic assays. When short synthetic nucleotides anneal to complementary sequences they can form a hybrid double stranded molecule capable of being recognised by polymerases, so targeting the polymerase to a specific sequence and thus location within the nucleic acid strand. This lecture will further discuss the theory and practice of PCR as a means of isolating, amplifying and manipulating DNA . It will then consider various applications of PCR and reverse transcription PCR (RT-PCR). These include the detection of RNA based pathogens such as viral influenza and HIV, the determination of viral or bacterial loads and forensic DNA fingerprinting using microsatellites or short tandem repeats. RT-PCR is also one of the most commonly used genomic tools, for the measurement of gene expression, providing us with a means of understanding and monitoring how the genome responds to disease, as well as its use in engineering and manipulation of DNA sequences in the development of modern therapeutics and vaccines. Learning Outcomes On successful completion of the lecture, students should be able to: Describe the principles of PCR amplification of DNA fragments Describe the reagents required for a PCR reaction to take place Describe how specificity of amplification is ensured Describe a range of modifications to the basic technique and what these are used for: Quantitative PCR (qPCR) SNP detection using High-resolution melting or qPCR Forensic use of microsatellite genetic markers (STRs)
-
What is Polymerase Chain Reaction (PCR)? (1)
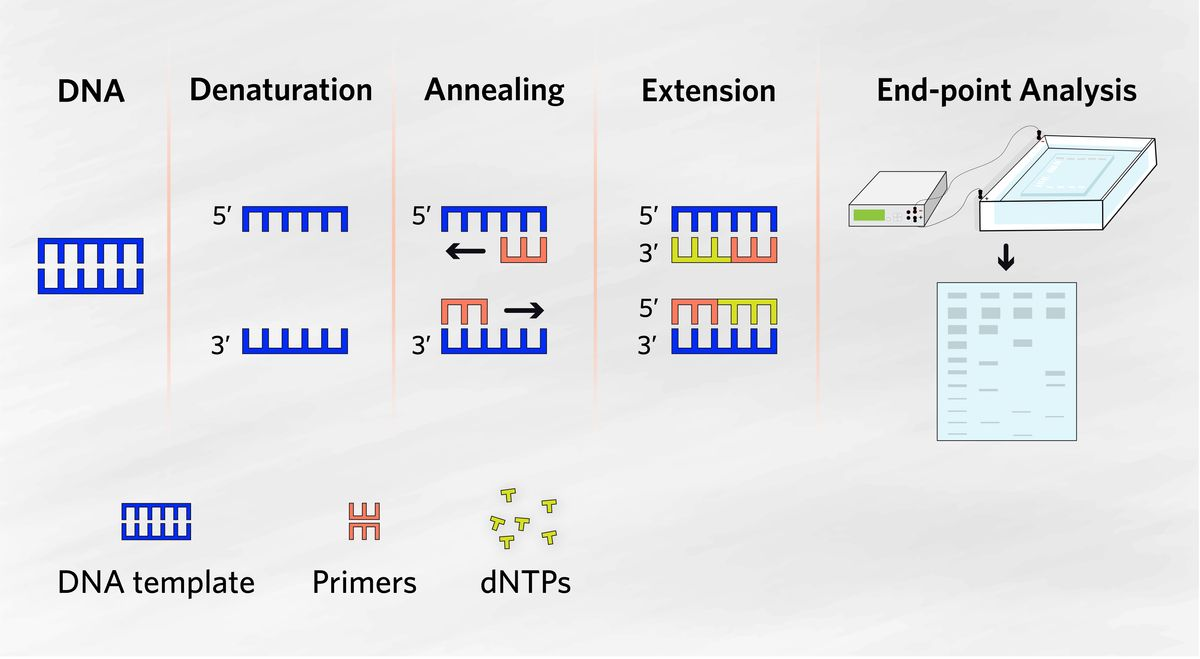
PCR is an enzyme-based method used to specifically amplify segments of DNA using a thermal DNA polymerase in a cyclical process.
-
What is a chain reaction in the context of PCR? (1)
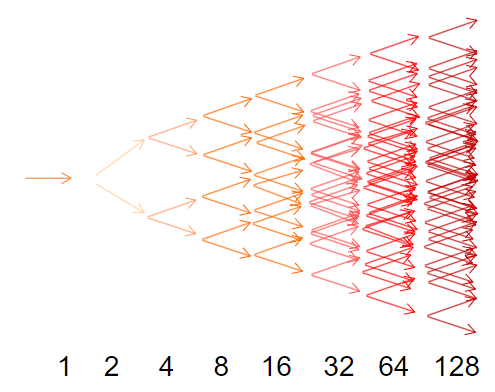
A chain reaction is a series of events, each dependent on the preceding event, leading to an exponential increase in the number of events occurring in sequence.
-
What is Polymerase Chain Reaction (PCR), and state its GENERAL importance? (3)
PCR is a method to specifically amplify segments of DNA.
It is specific only if annealing is undertaken at the melting temperature (Tm) of the primers, preventing mismatched base pairing.
Specificity stems from the complementarity of the primers.
-
What is Polymerase Chain Reaction (PCR), and state its importance (3)?
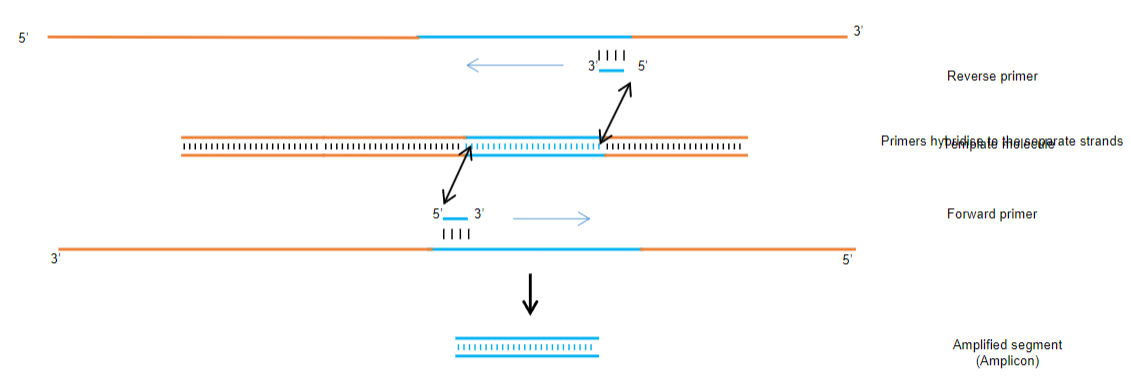
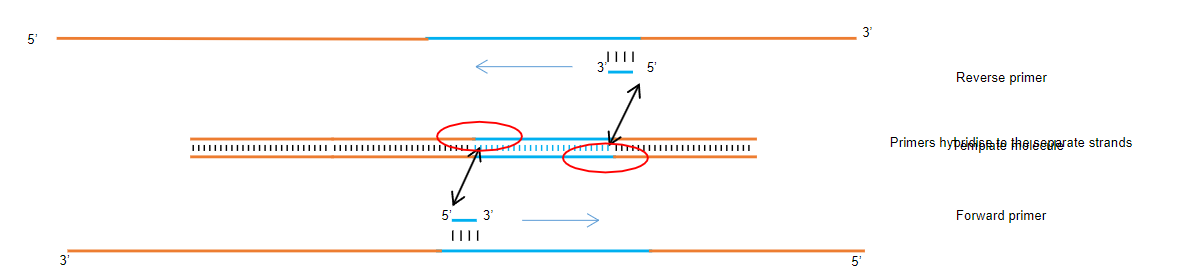
Specific Amplification: PCR allows the selective amplification of a specific DNA segment by using primers that are complementary to the target sequence at the ends.
Exponential Amplification: The process leads to an exponential increase in the number of copies of the target DNA segment, making it easier to analyse.
Utilization of Primers: Two primers are required, each complementary to the target sequence, ensuring that only the desired segment is amplified.
-
What is the role of DNA-dependent DNA polymerase in Polymerase Chain Reaction (PCR), and explain its function (2)?
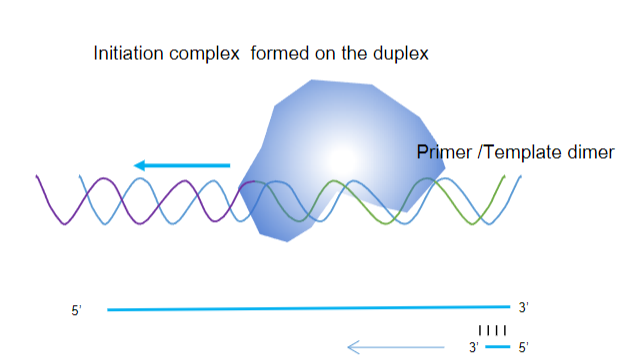
Recognizing the Initiation Complex: The enzyme recognizes a specific structure formed by the partially double-stranded DNA, allowing it to bind and initiate the reaction.
Extending the DNA Strand: It extends the newly synthesized strand from the 3’ end of the non-template strand, facilitating the amplification of the target DNA segment.
-
What is the significance of the primer/template duplex in Polymerase Chain Reaction (PCR), and describe its formation (3)?
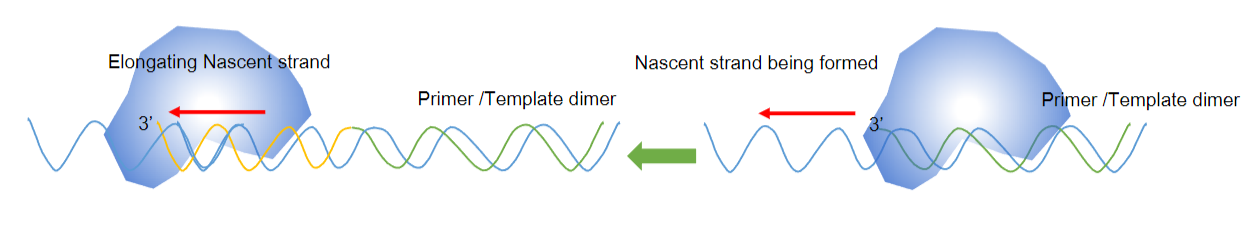
Answer: The primer/template duplex is significant in PCR because:
Denaturation of Template: The double-stranded template DNA must first be denatured into single strands by applying heat.
Formation of the Nascent Strand: The newly formed strand, known as the nascent strand, is synthesized as the PCR progresses.
Annealing of the Primer: A short single-stranded DNA molecule (the primer) anneals to the denatured template, forming a partially double-stranded structure that allows for DNA synthesis.
-
What occurs during the annealing of the primer to the template in Polymerase Chain Reaction (PCR), and why is it important (3)?
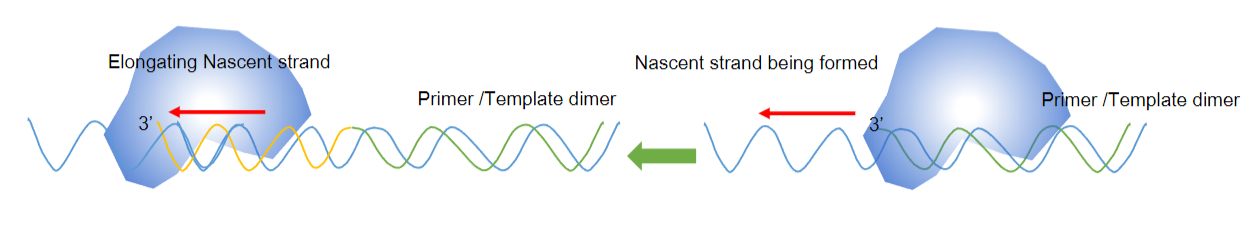
Answer: During the annealing of the primer to the template:
High Stringency Conditions: Annealing occurs under high stringency conditions, which are achieved by using the predicted melting temperature (Tm) of the primer-template duplex.
Base-Pairing Formation: This process is an alternative way of describing hybridization, where the primer binds to the template through complementary base-pairing.
Stabilization by Hydrogen Bonding: The annealed primer-template duplex is stabilized by hydrogen bonding, ensuring the specificity of the amplification process.
-
What is the difference between the annealing of primers and renaturation during Polymerase Chain Reaction (PCR), and what drives this preference (3)?
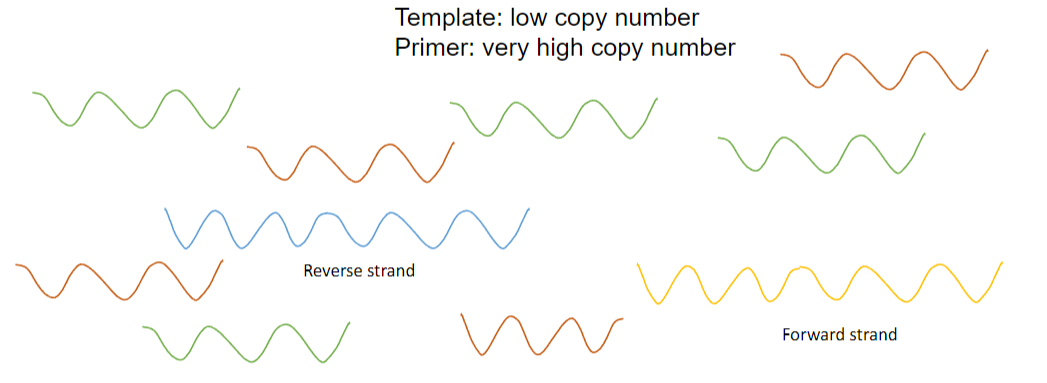
Answer: The difference between the annealing of primers and renaturation during PCR is that:
Competitive Process: Annealing of the primers and renaturation of the template are competitive processes, with primers present in very high copy numbers while templates are present in low copy numbers.
Preferential Annealing: The annealing of the primer occurs preferentially to renaturation because of the higher concentration of primers, which increases the likelihood of primer binding to the template.
Favourable Kinetics: This preference is driven by favourable kinetics, as the vast excess of the primer facilitates its rapid binding to the template over the slower renaturation of the template strands.
-
What are the basic rules of Polymerase Chain Reaction (PCR) regarding nucleic acid synthesis and the enzyme used (3)?
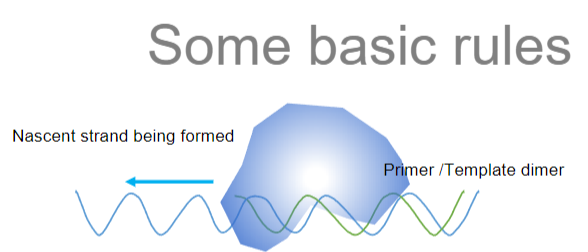
Answer: The basic rules of PCR regarding nucleic acid synthesis and the enzyme used are:
Synthesis of New Strands: PCR synthesizes a new nucleic acid strand by copying a DNA molecule.
Limitations with RNA: PCR cannot copy RNA nor make RNA; RNA must first be converted to DNA by reverse transcription before it can be amplified by PCR.
Enzyme Used: The enzyme used in PCR is a DNA-dependent DNA polymerase, which synthesizes new DNA strands from an existing DNA template.
-
What are the key requirements for a Polymerase Chain Reaction (PCR) to occur (4)?
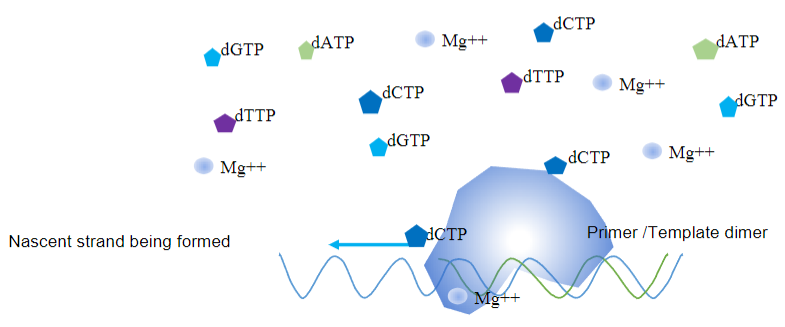
Answer: The key requirements for a PCR to occur are:
Template and Primers: A template with opposing primers (usually 20-30 bases long) annealed to the respective strands, with a free 3' OH and 5' overhangs.
Deoxynucleotide Triphosphates: Deoxy nucleotide triphosphates (dATP, dGTP, dCTP, dTTP) are needed for synthesizing new DNA strands.
Magnesium Ions: Mg²⁺ ions are required as a cofactor for the DNA polymerase enzyme.
Neutral pH: A roughly neutral pH environment is necessary for optimal enzyme activity.
-
What are the basic rules and states involved in the Polymerase Chain Reaction (PCR) process (3)?
Answer: The basic rules and states involved in the PCR process are:
Denatured State: The template becomes single-stranded due to heating, which breaks the hydrogen bonds between the DNA strands.
Annealed State: Primers hybridise to the single-stranded template, forming an initiating template for DNA synthesis.
Native State: The reaction reaches an optimal extension temperature and pH, allowing the DNA polymerase to synthesize new DNA strands efficiently.
-
What are the basic rules regarding thermostability and the importance of using thermostable polymerases in the Polymerase Chain Reaction (PCR) process (3)?

Answer: The basic rules regarding thermostability and polymerases in PCR are:
Multiple Rounds of Heating and Cooling: PCR requires several cycles of extreme heating and cooling to denature DNA and allow primer annealing, necessitating the use of thermostable enzymes.
Thermostability: Thermostable polymerases can retain their activity after being subjected to high temperatures that would typically denature other enzymes, ensuring successful DNA amplification.
Use of Thermophilic Polymerases: Enzymes like Taq polymerase, derived from thermophilic bacteria such as Thermus aquaticus, are commonly used in PCR due to their ability to withstand high temperatures during the denaturation step.
-
What are the typical cycling conditions in the Polymerase Chain Reaction (PCR) process, including denaturation, annealing, and extension temperatures, and their significance (4)?
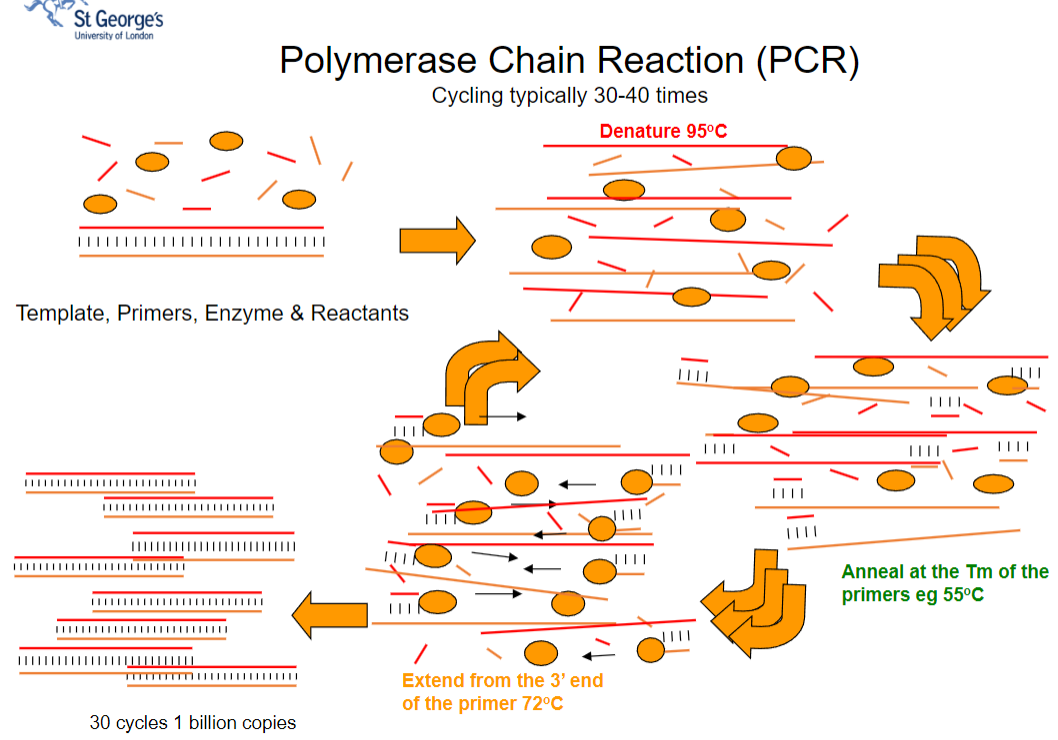
Answer: The typical cycling conditions in PCR include:
Denaturation (95°C): The template DNA is heated to separate its strands, allowing access for the primers.
Annealing (around 55°C): Primers hybridize to the single-stranded template at their melting temperature (Tm), ensuring specificity in the amplification process.
Extension (72°C): DNA polymerase extends the primers from their 3' ends, synthesizing new DNA strands.
Cycling: This process is typically repeated 30-40 times, resulting in approximately 1 billion copies of the target DNA sequence.
-
What are the key characteristics of the accumulation of product in Polymerase Chain Reaction (PCR) and the factors influencing its kinetics (3)?
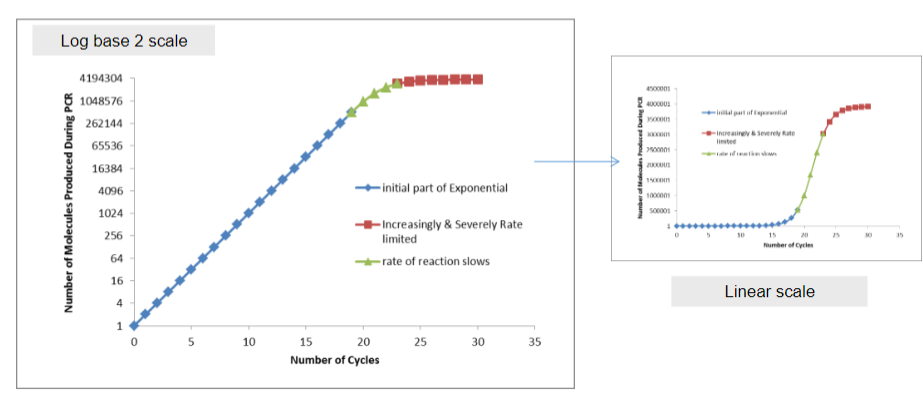
Answer: The key characteristics of product accumulation in PCR include:
Exponential Accumulation: Every cycle of PCR results in a doubling of the amount of product, leading to exponential growth of the target DNA.
Reaction Kinetics: The kinetics of the reaction are influenced by the depletion of reactants, which can slow down the reaction as the components become less available.
Acidification: The accumulation of products can lead to acidification of the reaction mixture, which may further impact enzyme activity and reaction efficiency.
-
What are the applications of Polymerase Chain Reaction (PCR) in molecular biology and clinical diagnostics (4)?
Answer: The applications of PCR in molecular biology and clinical diagnostics include:
Routine Diagnostics: PCR serves as a standard diagnostic tool for identifying, confirming, and quantifying specific DNA sequences.
Presence/Absence Testing: It is used for detecting the presence or absence of pathogens, such as identifying tuberculosis (TB) in sputum samples and determining appropriate treatment choices.
Differentiation of Organisms: PCR can differentiate between closely related organisms, for example, distinguishing between swine flu and human influenza, specifically within the H1N1 subtype.
Viral Load Measurement: PCR helps determine the quantity of viral RNA, such as measuring the HIV viral load to guide treatment decisions and monitoring the presence of SARS-CoV-2 in individuals for COVID-19 diagnosis.
-
What is the limitation of the standard Polymerase Chain Reaction (PCR) in terms of output, and how is this limitation addressed in real-time PCR (2)?
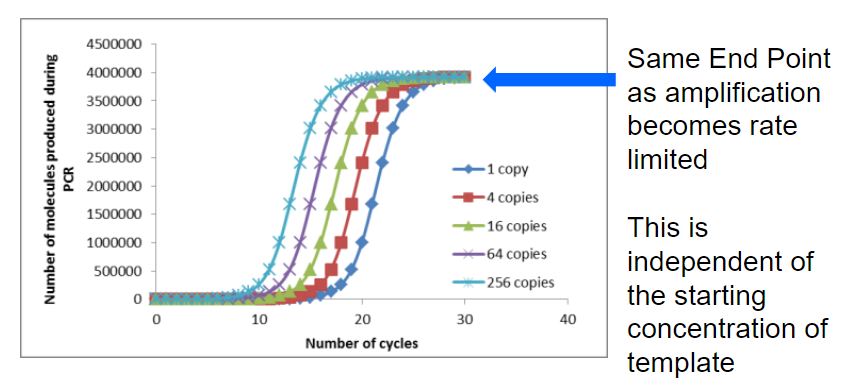
Answer: The limitation of standard PCR is that it does not provide a quantitative output and cannot determine the template copy number at the end of the reaction. This limitation is addressed in real-time PCR, which involves modifications to the standard technique that allow for measurable output during the exponential phase of amplification, enabling the quantification of DNA in real-time.
-
What are the features of quantitative PCR (qPCR), and how is it used in diagnostics? (3)
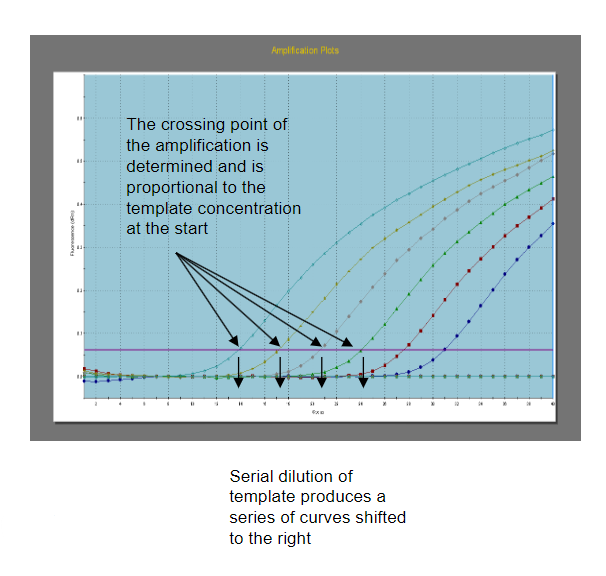
Answer: Quantitative PCR (qPCR), also known as real-time PCR, utilizes fluorescent detection methods to monitor the amplification of target DNA molecules during the PCR process. It is used in diagnostics for quantifying the amount of a specific target DNA molecule in a sample, enabling accurate assessment of DNA levels.
-
What are single nucleotide polymorphisms (SNPs), and what are their common applications in PCR? (3)
Answer: Single nucleotide polymorphisms (SNPs) are single nucleotide genetic variants that can affect an individual's susceptibility to diseases, treatment responses, and drug sensitivity.
Common applications of SNP detection in PCR include antibiotic resistance testing (e.g., for TB), identification of genetic markers related to drug sensitivity (e.g., CYP2C9 and VKORC1 variants for warfarin sensitivity), and markers for diseases like cancer.
Several methodologies, including adaptations of quantitative real-time PCR, are used to detect these genetic variants based on differences in the melting temperature (Tm) of short DNA sequences.
-
What are the two approaches used for single nucleotide polymorphism (SNP) detection in PCR, and how do they work? (2)
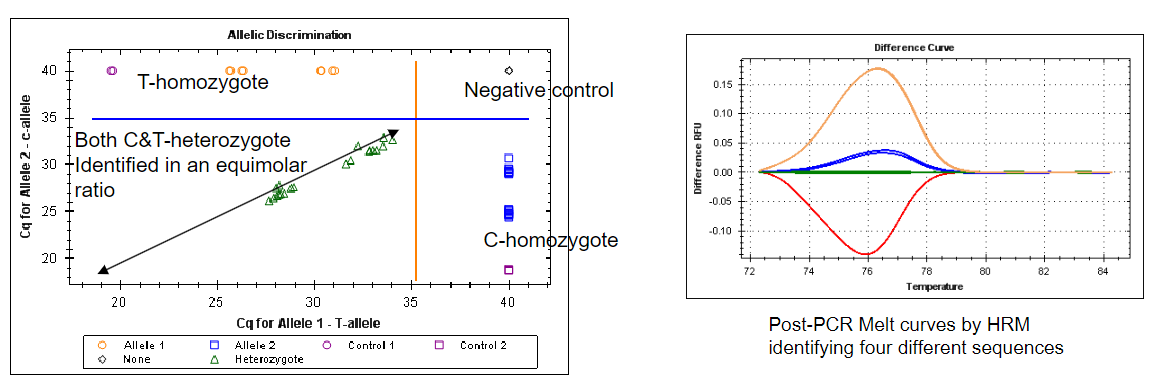
Answer: The two approaches used for single nucleotide polymorphism (SNP) detection in PCR are:
High Resolution Melting (HRM): This method uses the melting temperature (Tm) of the amplified product to determine which sequence variant is present. Differences in Tm can indicate the presence of specific SNPs.
Probe-Based Quantitative PCR (qPCR): Sometimes referred to as allelic discrimination, this approach involves the specific binding of a probe to the amplified region containing the SNP. The detection of this binding allows for the identification of the specific variant present in the sample.
-
What are the applications of PCR in forensics and law enforcement? (4)
Answer: The applications of PCR in forensics and law enforcement include:
Parentage or Kinship Testing: Used for immigration purposes and inheritance disputes.
Identification: Helps identify military casualties, missing persons, or victims of environmental disasters.
Matching Biological Materials: Used to match biological materials from two sources, aiding in placing an individual at a crime scene.
Authentication of Biological Material: Ensures the authenticity of cell lines and the purity of food products.
-
What are short tandem repeats (STRs) and their role in forensic identification? (4)
Answer: Short tandem repeats (STRs) are repetitive sequences in the genome used for forensic identification. They consist of 2-5 or more bases repeated many times at specific locations. Key points include:
High Polymorphism: The number of repeats varies between individuals, making STRs highly polymorphic.
Unique Patterns: Each individual's genome provides a pattern of uniquely sized products based on their STRs.
Location Diversity: Many different STRs are scattered throughout the genome.
Forensic Application: STRs are utilized in forensic identification to distinguish between individuals based on their genetic profiles.
-
What is the significance of short tandem repeats (STRs) in forensic analysis, particularly regarding the UK DNA database? (4)
Answer: Short tandem repeats (STRs) play a crucial role in forensic analysis, especially in the UK DNA database. Key points include:
Current Database Composition: The UK DNA database currently consists of 10 STRs used for identification.
Molecular Bar Code: STRs provide something akin to a molecular bar code or "DNA fingerprint" for individuals.
Size Variation: Each STR differs in size, resulting in a total of 20 numbers and a gender indicator for identification purposes.
Matching Probability: Together, these STRs offer a matching probability with an error of around 1 in 1 billion, enhancing the reliability of forensic identification.
-
How do multiple sets of labeled primers enhance the uniqueness of DNA fingerprints in STR analysis? (2)
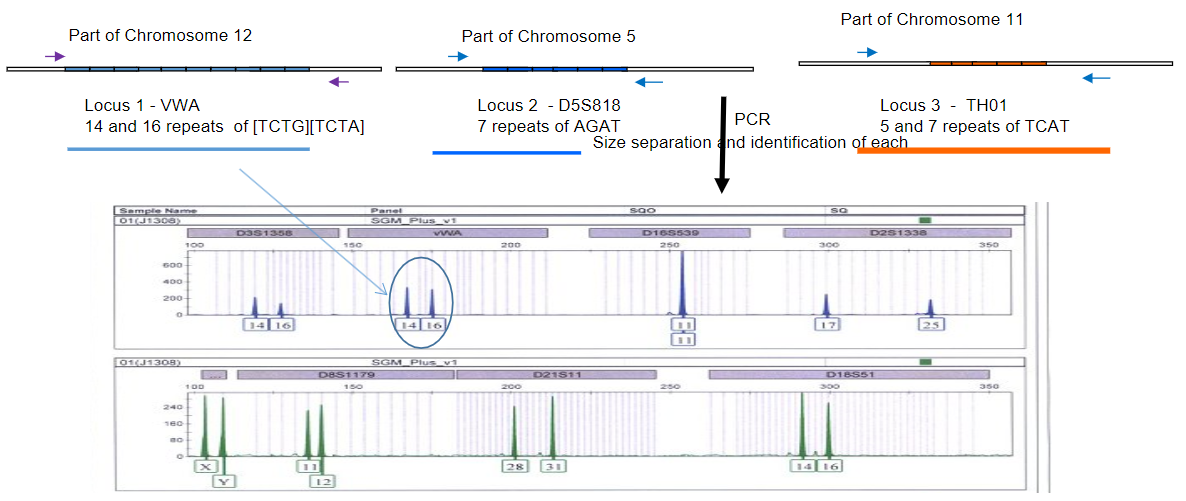
Answer: Multiple sets of labeled primers enhance the uniqueness of DNA fingerprints in STR analysis by:
Targeting Different STRs: Each set of primers is designed to amplify different short tandem repeats (STRs) across the genome, resulting in a variety of amplified products.
Creating Unique Size Patterns: By investigating more STRs, a more unique pattern of sizes is produced, contributing to a distinct "DNA fingerprint" for individuals, which improves identification accuracy.
-
Picture demonstrating PCR STRs
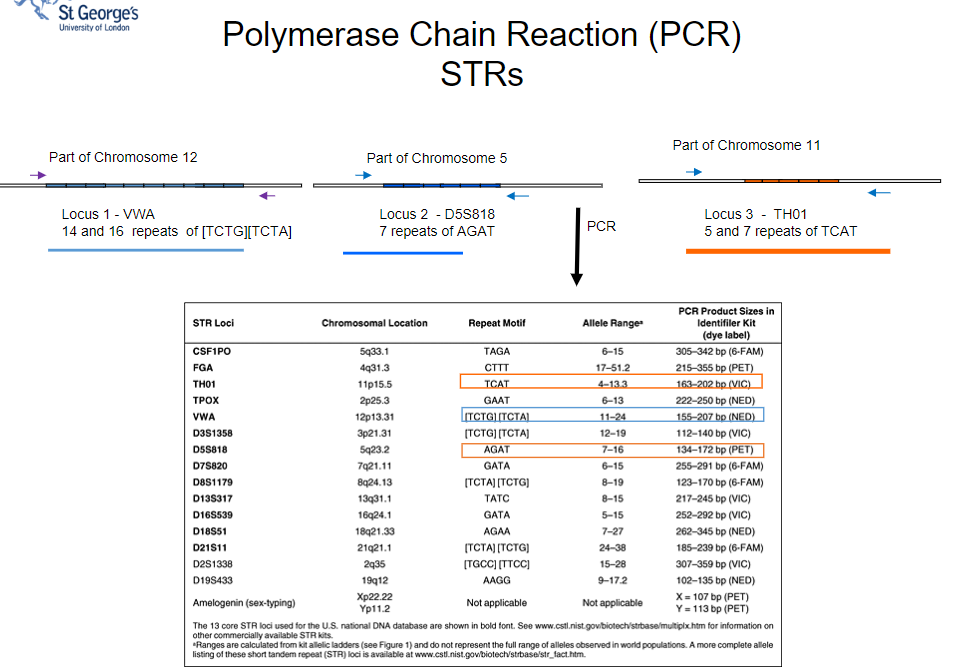
-
What are the other applications of Polymerase Chain Reaction (PCR)? (5)
Answer: The other applications of Polymerase Chain Reaction (PCR) include:
Amplifying Material for Next Generation Sequencing: This involves simultaneously sequencing a large number of PCR products of candidate cancer genes.
Isolating DNA Segments: PCR is used to isolate individual segments of DNA before cloning or sequencing.
Manipulating and Modifying DNA: It can introduce mutations into a sequence of DNA.
Modifying DNA Ends: PCR modifies the ends of a DNA sequence to create restriction sites compatible with cloning vectors.
Recombinant DNA Technology: PCR is crucial for developing recombinant vaccines and pharmaceuticals, such as interferons, clotting factors, and tissue plasminogen activator.
-
What are the key points discussed in the summary of Polymerase Chain Reaction (PCR)? (6)
Answer: The key points discussed in the summary of Polymerase Chain Reaction (PCR) include:
The main modifications of PCR that enable it to be quantitative.
The use of PCR as a major diagnostic tool, specifically real-time PCR, high-resolution melting, and allelic discrimination.
Its application in detecting and quantifying specific organisms, such as infectious agents.
The identification of recombination events in cancer, specifically BCR-ABL.
The detection and confirmation of single nucleotide polymorphisms (SNPs) or mutations in gene sequences.
The amplification of short tandem repeats (STRs) for forensic applications and its role in manipulating and modifying DNA as a vital tool in recombinant DNA technology.

