Mapping Mendelian Disease (Genomics)
Mapping Mendelian Disease In this session, Prof. Pia Ostergaard will share some of her research into the genetics of primary lymphoedema, as an exemplar of gene identification in rare genetic disease. Before the live session, please make sure to review the pre-lecture refreshers in Sections 1 and 2 below. Learning Outcomes On successful completion of the lecture, students should be able to: Describe the principles of linkage analysis Describe the lymphatic system and what primary lymphoedema is Describe how linkage analysis has been successfully applied to primary lymphoedema to identify causative gene variants in conjunction with Sanger sequencing and Next Generation Sequencing
-
Why is recombination important? (3)
Recombination creates genetic diversity by shuffling alleles between homologous chromosomes.
It ensures new combinations of traits in offspring, which is vital for evolution.
Helps repair DNA damage and maintain chromosome integrity.
-
What is an allele? (2)
An allele is a variant form of a gene at a specific locus on a chromosome.
Different alleles can result in variations in traits, such as eye colour or blood type.
-
If two genes are on the same chromosome, are they always inherited together? (2)
No, they may be separated during recombination in meiosis.
The likelihood depends on their distance on the chromosome—closer genes are less likely to recombine.
-
If two alleles are on the same chromosome, are they always linked and inherited together? (2)
Not always; linkage depends on their proximity.
Recombination events can separate linked alleles if they are far enough apart on the chromosome.
-
What does the lymphatic system look like? (3)
A network of thin, vein-like vessels spread throughout the body.
Includes lymph nodes, small bean-shaped structures located along these vessels.
Contains organs like the spleen, thymus, and tonsils that support immune function.
-
What is one of the roles of the lymphatic system? (3)
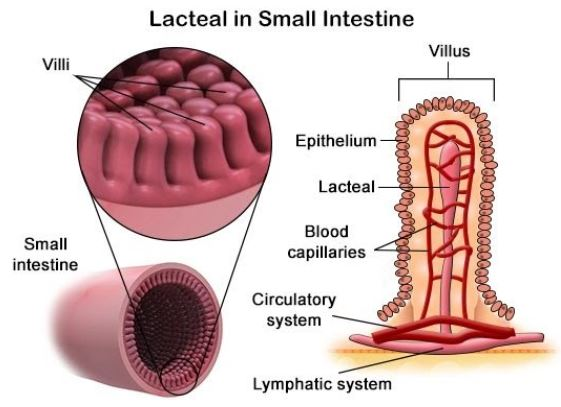
Maintains fluid balance by collecting excess fluid and returning it to the bloodstream.
Plays a key role in immune defense by transporting white blood cells and filtering pathogens.
Absorbs fats and fat-soluble vitamins from the digestive system via the lacteals.
-
What is lymphoedema? (2)
A condition caused by the buildup of lymph fluid, leading to swelling, often in the arms or legs.
Results from damage or obstruction in the lymphatic system, such as from surgery or infection.
-
How is lymphodema treated? (4)
Compression therapy: Using bandages or garments to improve fluid drainage.
Manual lymphatic drainage: A specialized massage technique to encourage fluid movement.
Exercise: Promotes circulation and lymphatic flow.
In severe cases, surgery may be performed to remove excess tissue or improve lymphatic drainage.
-
Why are lymphatics called the "Cinderella of the circulatory system"? (1)
They are often overlooked compared to the blood circulatory system but play an integral role in health.
-
What are the functions of the lymphatic system? (3)
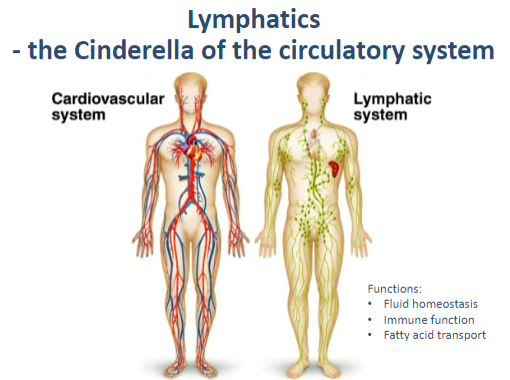
Fluid homeostasis: Maintains balance by returning excess interstitial fluid to the bloodstream.
Immune function: Filters pathogens and supports immune responses.
Fatty acid transport: Absorbs fats and fat-soluble vitamins from the digestive tract.
-
How are lymphatics part of the circulatory network? (1)
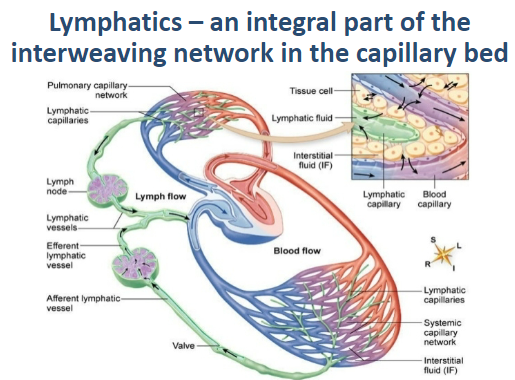
They form an interweaving network in the capillary bed, working closely with blood vessels.
-
What is primary lymphoedema? (3)
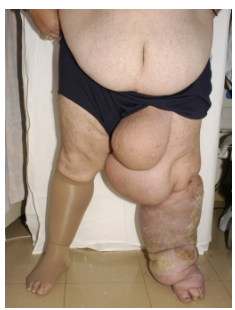
Chronic oedema caused by a developmental abnormality or dysfunction of the lymphatic system.
Often progressive, with phenotypes varying by age of onset, site, inheritance patterns, associated features, and genetic causes.
Leads to debilitating, embarrassing, and stressful symptoms with recurrent infections.
-
Why is research on lymphoedema important? (2)
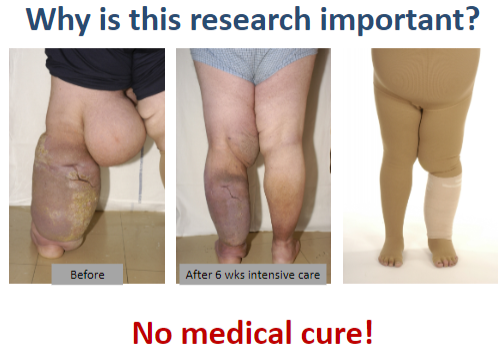
There is currently no medical cure for lymphoedema.
Research bridges the gap between laboratory findings ("bench") and practical patient treatments ("bedside").
-
How do we study genetic diseases related to the lymphatic system? (1)
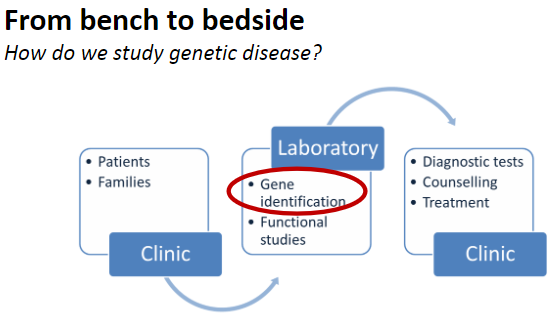
By investigating genetic causes, inheritance patterns, and phenotypes using modern genetic and molecular techniques.
-
What are the steps in studying genetic diseases? (3)
Gene identification by gene mapping:
Homozygosity mapping
Linkage analysis
GWAS (Genome-Wide Association Studies)
Finding disease-causing mutations:
Sequencing
Proving causation:
Using in silico, in vitro, and in vivo tools.
-
What is genetic linkage? (3)
The tendency for alleles at neighbouring loci (e.g., A1, A2, B1, and B2) to segregate together during meiosis.
To be linked, two loci must lie very close together on the chromosome.
Linked loci are less likely to be separated by recombination during meiosis.
-
What is a haplotype, and how is it useful? (2)
A haplotype defines multiple alleles at linked loci, marking chromosomal segments.
These segments can be tracked through pedigrees and populations to study inheritance patterns.
-
How does recombination affect genetic linkage? (2)
Recombination (cross-overs) during meiosis is more likely between loci separated by a greater distance.
Closely linked loci are less likely to be separated, maintaining their association across generations.
-
Why is understanding genetic linkage important for gene mapping? (1)
It helps identify loci associated with diseases by tracking how linked alleles segregate together in pedigrees and populations.
-
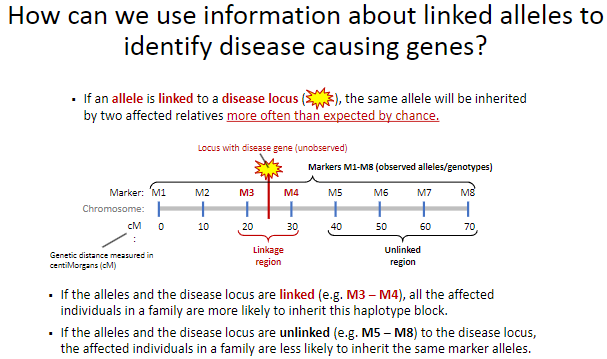
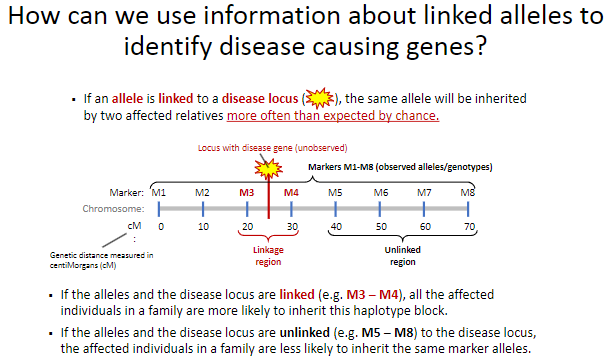
How can linked alleles help identify disease-causing genes? (2)
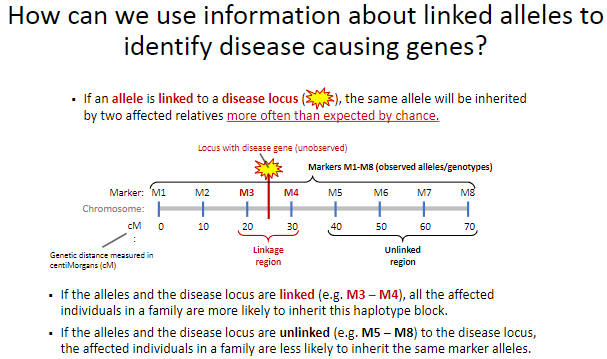
If an allele is linked to a disease locus, affected relatives will inherit the same allele more often than expected by chance.
This linkage helps trace the disease locus through families by identifying shared haplotype blocks.
-
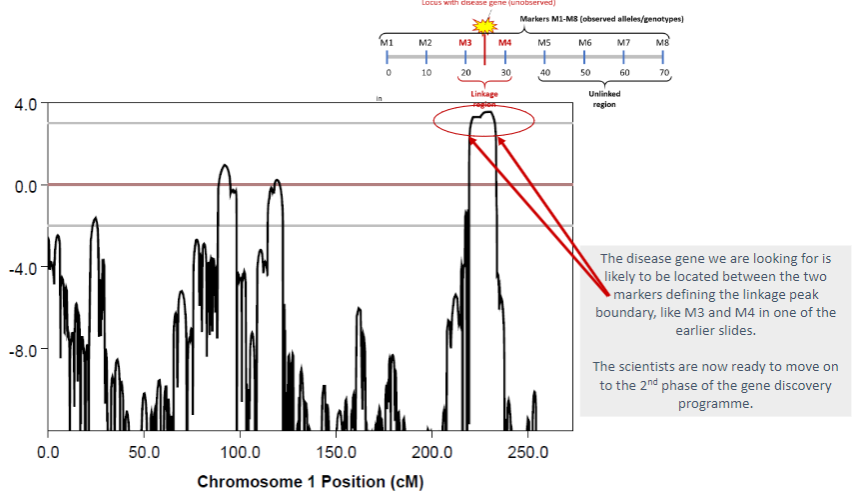
What happens when alleles are linked to a disease locus? (2)
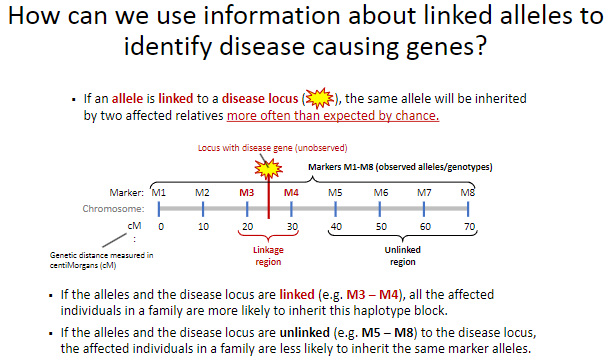
Affected individuals in a family are more likely to inherit the same haplotype block (e.g., M3 – M4) as the disease locus.
This shared inheritance pattern indicates the proximity of the alleles to the disease locus.
-
What happens when alleles are unlinked to a disease locus? (2)
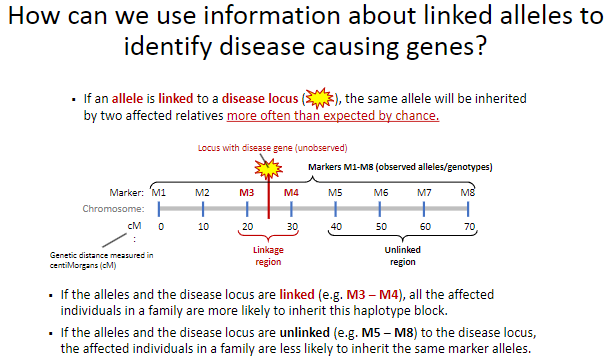
Affected individuals in a family are less likely to inherit the same marker alleles (e.g., M5 – M8).
This suggests that the alleles and the disease locus are located far apart or on different chromosomes.
-
Why is haplotype inheritance significant in linkage studies? (1)
Haplotype blocks mark chromosomal regions and can indicate the genomic location of the disease-causing gene.
-
What is linkage analysis? (2)
A gene mapping tool used to identify the genomic regions linked to diseases.
It involves using observed loci (alleles) to infer the location of unobserved loci (such as disease genes).
-
What type of design is used in linkage analysis? (1)
Family-based design, ranging from using a few large families to many small nuclear families or sib-pairs.
-
What is the goal of linkage analysis? (1)
The primary goal is to find the genomic region(s) that are linked to a disease.
-
How is linkage analysis used in the study of primary lymphoedema? (2)
Traditional linkage analysis and Sanger sequencing are used to identify the cause of autosomal recessive primary lymphoedema.
These methods help pinpoint the genetic loci linked to the disease and identify the mutations responsible for the condition.
-
What genetic inheritance pattern is associated with primary lymphoedema? (1)
Primary lymphoedema often follows an autosomal recessive inheritance pattern.
-
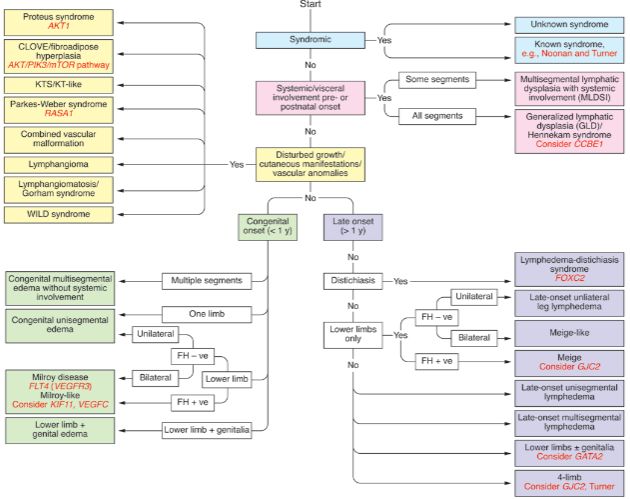
What is generalized lymphatic dysplasia? (1)
Generalized lymphatic dysplasia refers to a condition characterized by abnormal lymphatic development, leading to fluid buildup and edema in various parts of the body.
-
What are the main features of Hennekam syndrome (HS)? (5)
Antenatal hydrops with ascites and pleural effusions
Oedematous at birth
Intestinal lymphangiectasia
Peripheral lymphoedema affecting the arms, legs, and face
Mild developmental delay
-
What is the first step in performing a linkage analysis? (1)
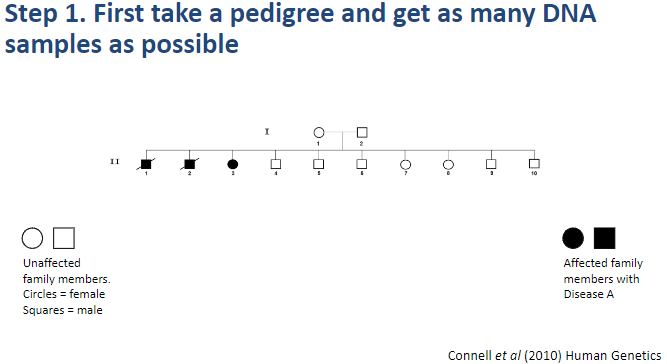
Step 1: Take a pedigree and collect as many DNA samples as possible.
-
What is the second step in performing a linkage analysis? (1)
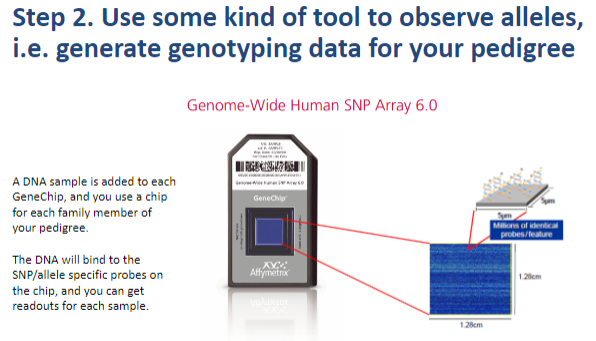
Step 2: Use a tool to observe alleles and generate genotyping data for the pedigree.
-
What is the third step in performing a linkage analysis? (1)
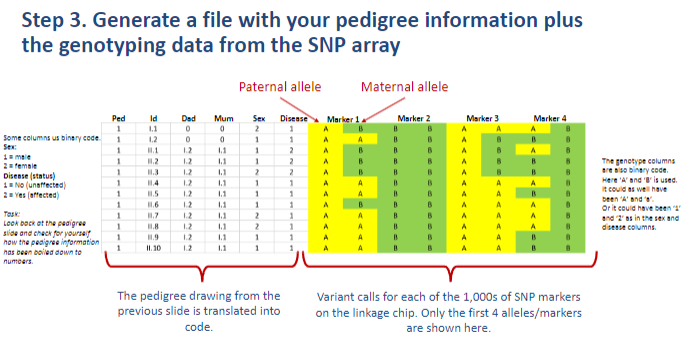
Step 3: Generate a file with the pedigree information along with genotyping data from the SNP array.
-
What is the fourth step in performing a linkage analysis? (1)
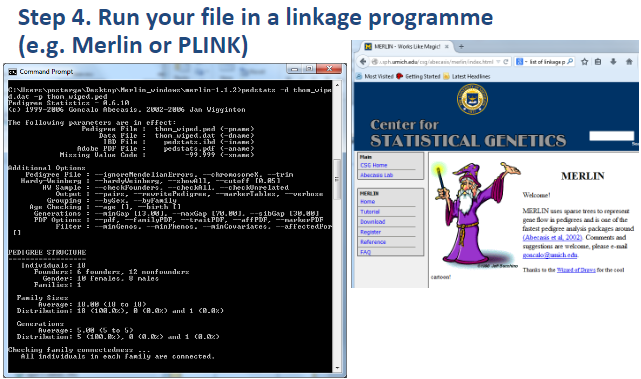
Step 4: Run the file in a linkage program (e.g., Merlin or PLINK).
-
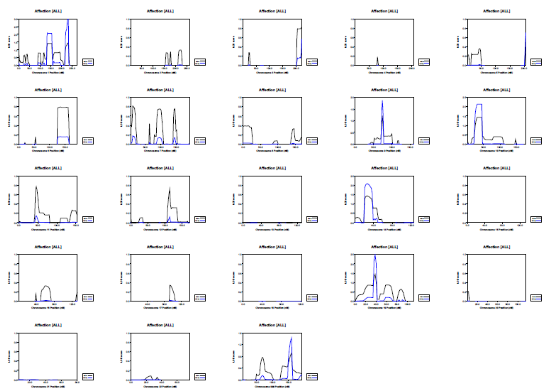
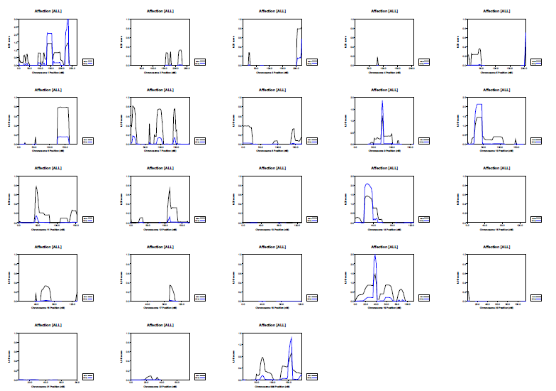
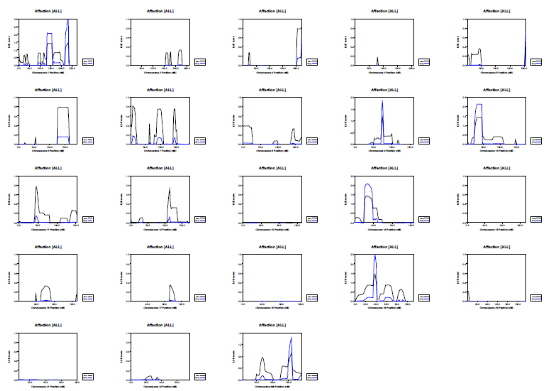
Picture demonstrating Example of the genetic distribution of markers (alleles) on a genotyping SNP array:
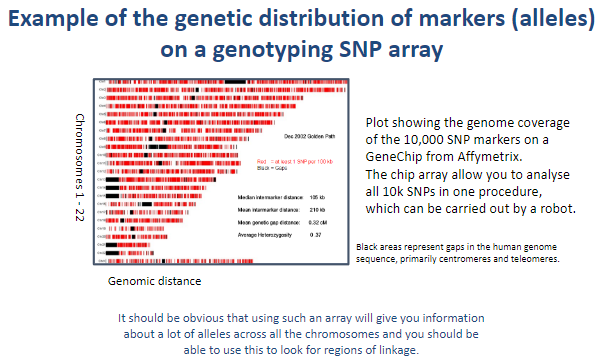
-
What does NPL stand for in genetic studies? (1)
NPL stands for Nonparametric Linkage.
-
What does nonparametric linkage testing (NPL) provide in genetic studies? (1)
NPL provides a plot for each chromosome to assess linkage.
-
How does NPL differ from parametric linkage testing? (1)
NPL does not require assumptions about the inheritance model (no need to specify a genetic model or pedigree structure), unlike parametric linkage testing.
-
What is a key characteristic of nonparametric linkage (NPL) testing? (1)
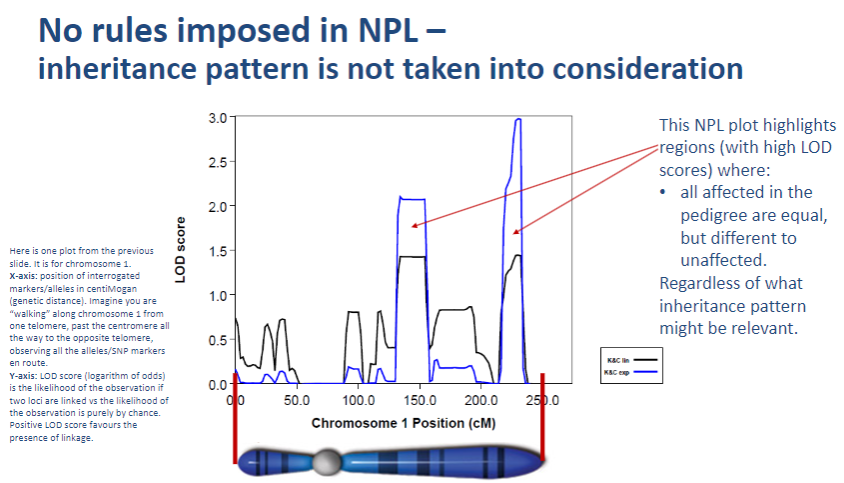
NPL does not impose any rules on the inheritance pattern, meaning it does not require assumptions about the genetic model.
-
Why doesn't NPL take inheritance patterns into account? (1)
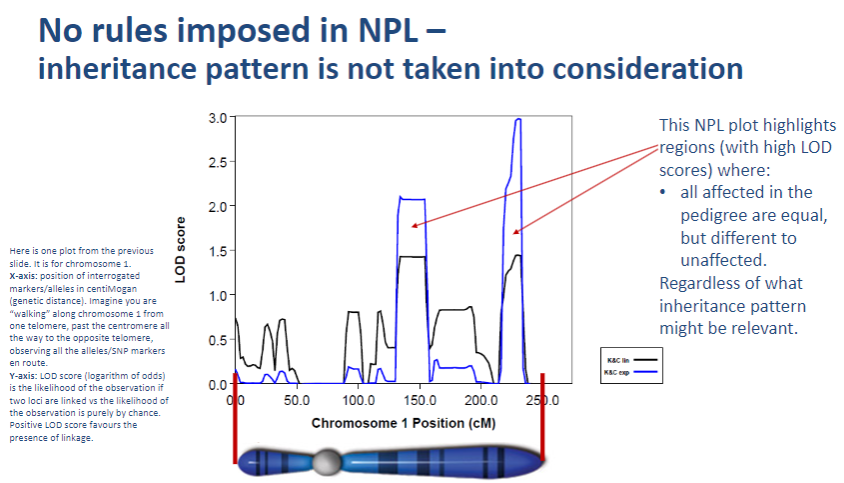
NPL focuses on detecting linkage by observing allele sharing between affected individuals without needing a predefined inheritance model.
-
How does NPL assess genetic linkage? (1)
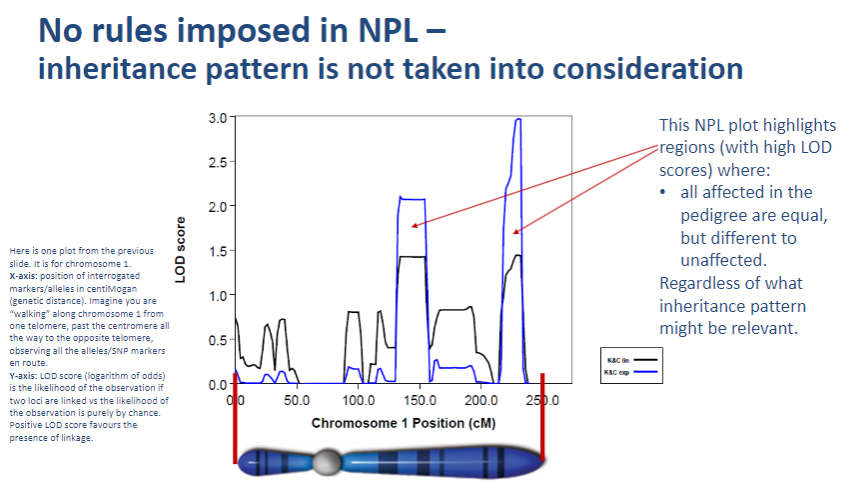
NPL assesses genetic linkage by comparing allele patterns across families and identifying whether certain alleles are shared more frequently in affected individuals.
-
What does parametric analysis impose in genetic studies? (1)
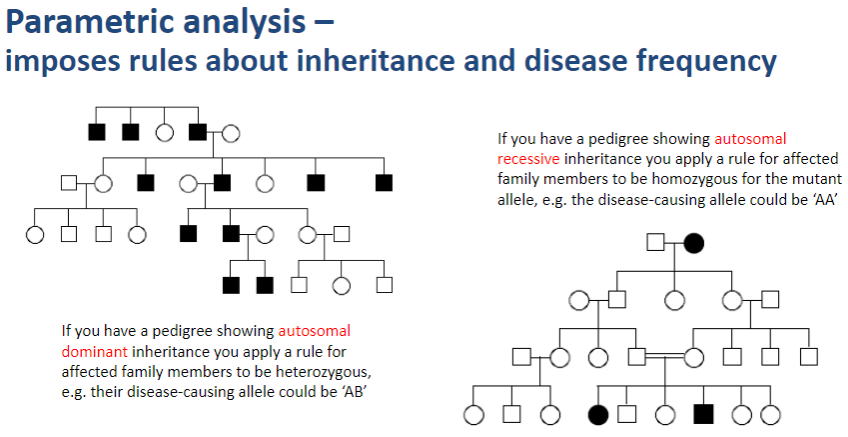
Parametric analysis imposes rules about inheritance patterns and disease frequency when assessing genetic linkage.
-
How does parametric analysis apply inheritance rules? (2)
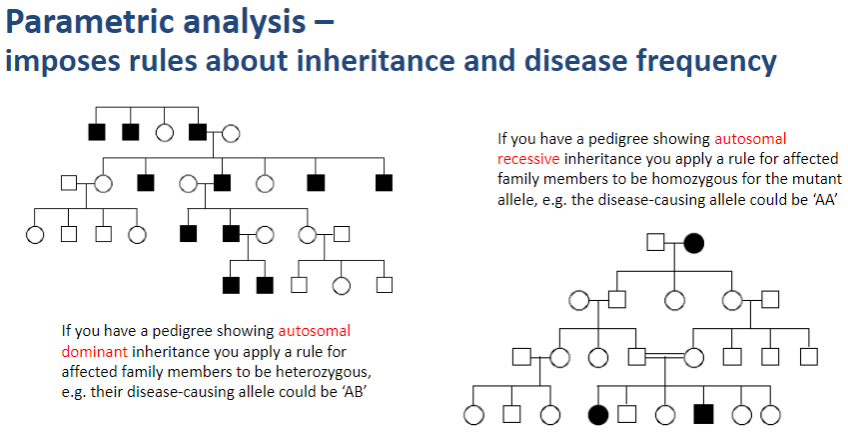
In autosomal dominant inheritance, affected family members are assumed to be heterozygous for the disease-causing allele (e.g., AB).
In autosomal recessive inheritance, affected family members are assumed to be homozygous for the mutant allele (e.g., AA).
-
Why is parametric analysis important in gene mapping? (1)
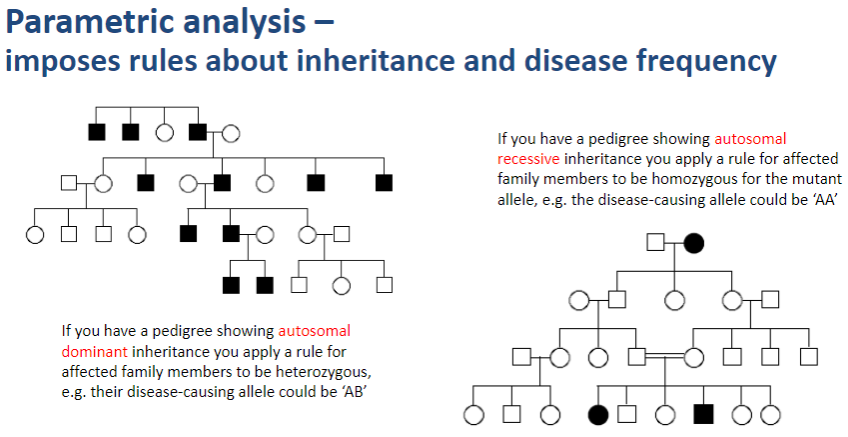
Parametric analysis helps refine linkage results by taking into account specific inheritance patterns and disease allele frequencies, improving the accuracy of mapping disease-causing genes.
-
What does a parametric test display in linkage analysis? (2)
The result is displayed for each of the 23 chromosomes.
Parametric tests highlight regions with high LOD scores, indicating areas where affected individuals show a distinct inheritance pattern compared to unaffected individuals.
-
What do high LOD scores indicate in parametric linkage analysis? (2)
High LOD scores indicate regions where the affected individuals' genotypes follow the imposed inheritance pattern, and differ from unaffected individuals.
These regions are potential candidates for being linked to a disease-causing gene.
-
How does parametric analysis help in interpreting genotyping data? (1)
Parametric analysis helps by comparing the genotypes of affected vs. unaffected individuals, ensuring that the genotyping follows the expected inheritance pattern for the disease under study.
-
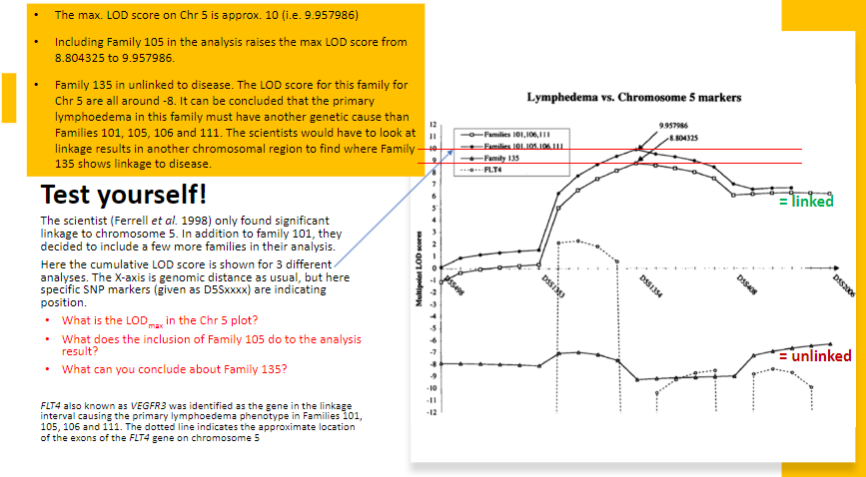
What does a LOD score greater than 3.0 indicate in linkage analysis? (1)
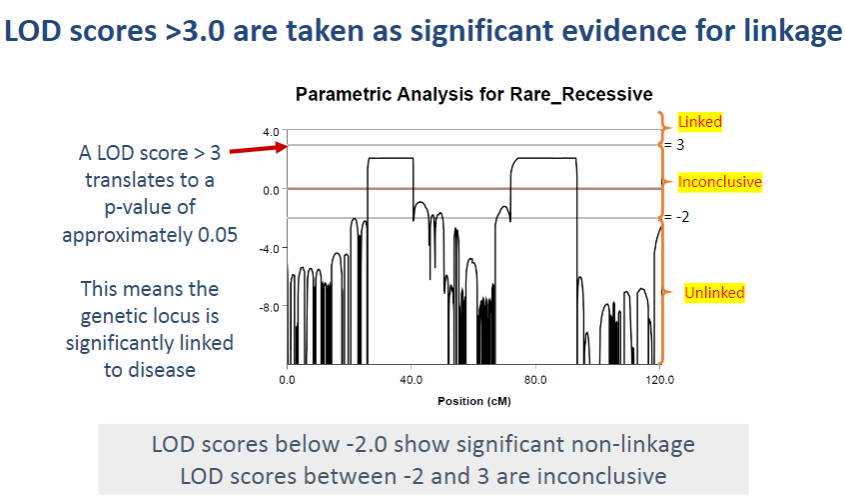
A LOD score > 3.0 is considered significant evidence for linkage, meaning the genetic locus is likely linked to the disease.
-
What does a LOD score of 3 translate to in terms of statistical significance? (1)
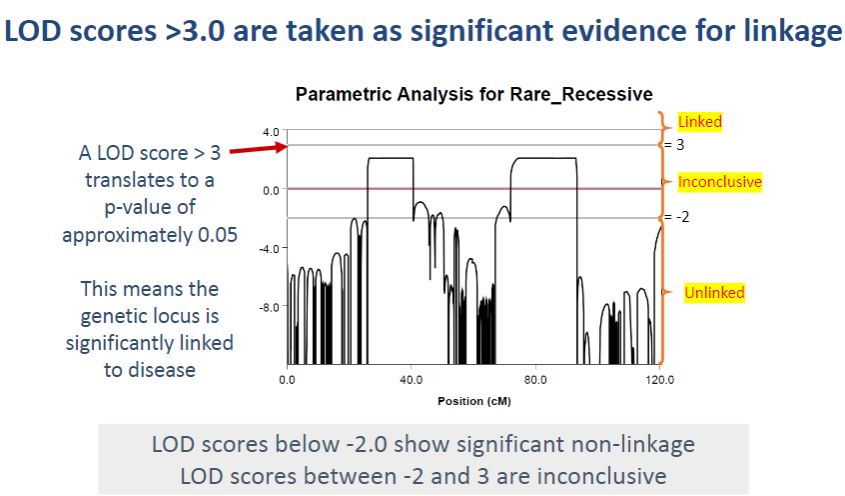
A LOD score > 3 corresponds to a p-value of approximately 0.05, which indicates statistical significance in the association between the genetic locus and the disease.
-
What do LOD scores between -2 and 3 indicate? (1)
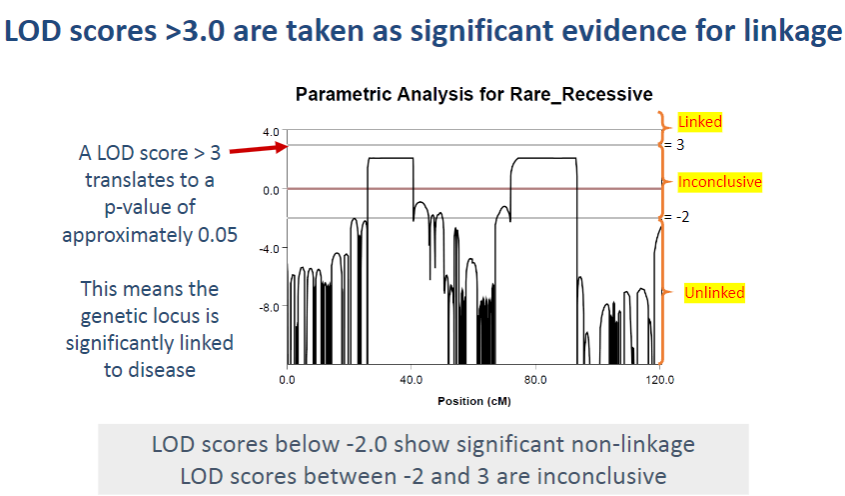
LOD scores between -2 and 3 are considered inconclusive, as they do not provide strong evidence for or against linkage.
-
What do LOD scores below -2 indicate? (1)
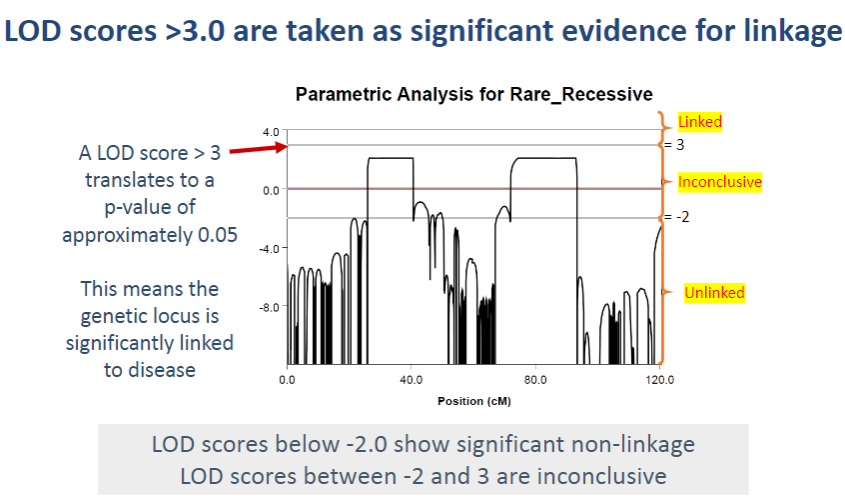
LOD scores below -2 suggest significant non-linkage, meaning the genetic locus is unlikely to be associated with the disease.
-
What could be the value of exploring regions with a LOD score between -2 and 2.1 in this analysis? (2)
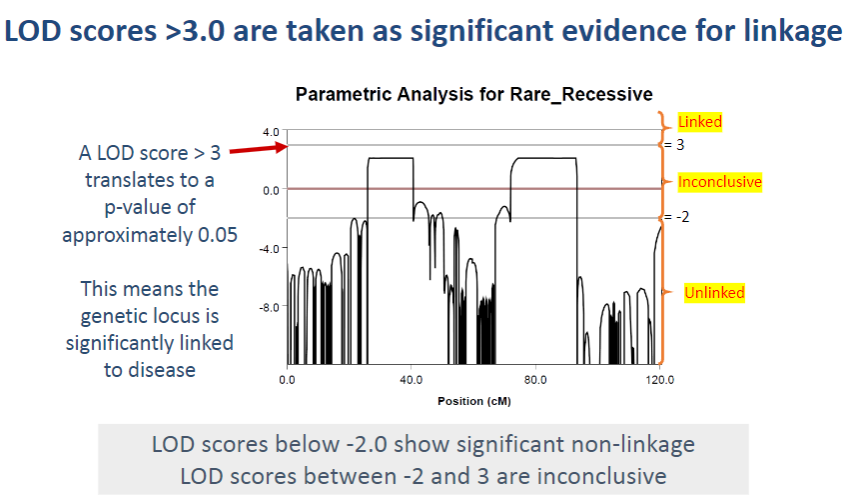
Even though a LODmax of 2.1 is not significant, the two regions with LOD scores between -2 and 2.1 are worth exploring further because they were the only two regions above LOD score -2. This suggests they may be linked to the disease, but more investigation is needed.
-
Why would the regions with inconclusive results be considered for further exploration? (1)
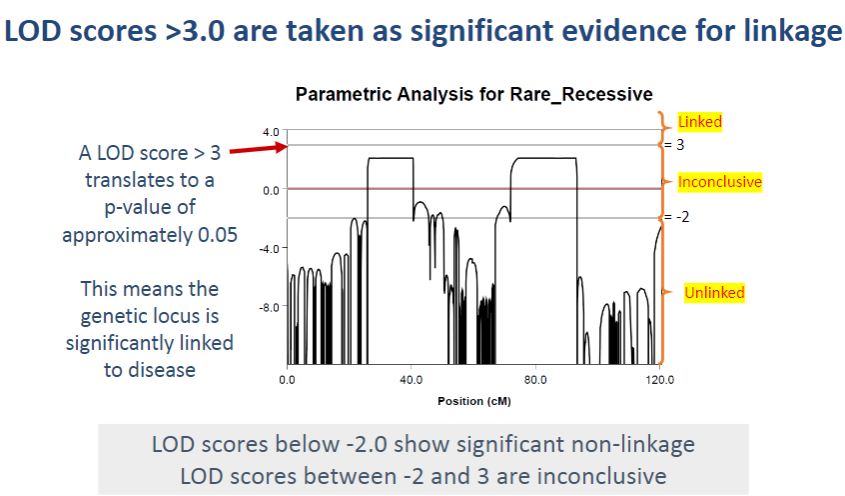
These regions are considered for further exploration because they may still have a genetic association with the disease, especially as they were the only regions that exceeded the -2 LOD score threshold when applying parametric analysis for autosomal recessive inheritance.
-
Picture demonstrating Result from linkage analysis of HS family:‘Linkage’ only to chromosome 18:
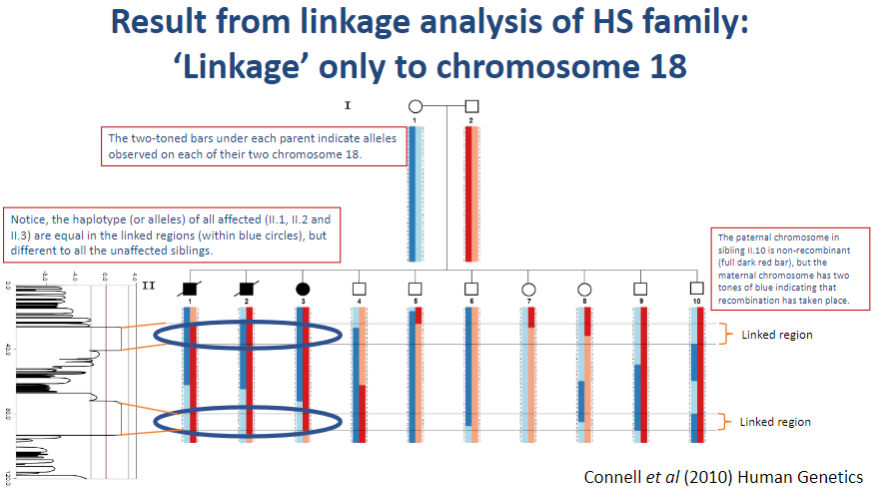
-
What is the first step in identifying disease-causing mutations? (1)
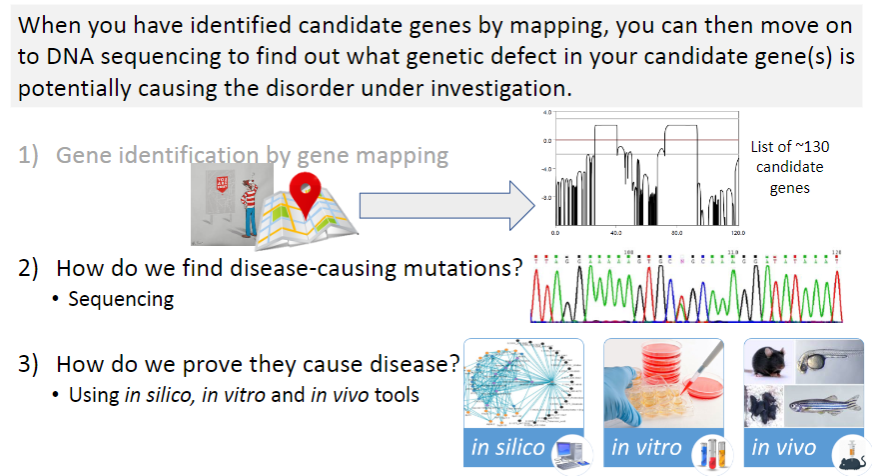
Gene identification by gene mapping: This step involves identifying regions of the genome that are linked to a disease through methods like linkage analysis and GWAS.
-
Once candidate genes are identified, what method is used to find the specific disease-causing mutation? (1)
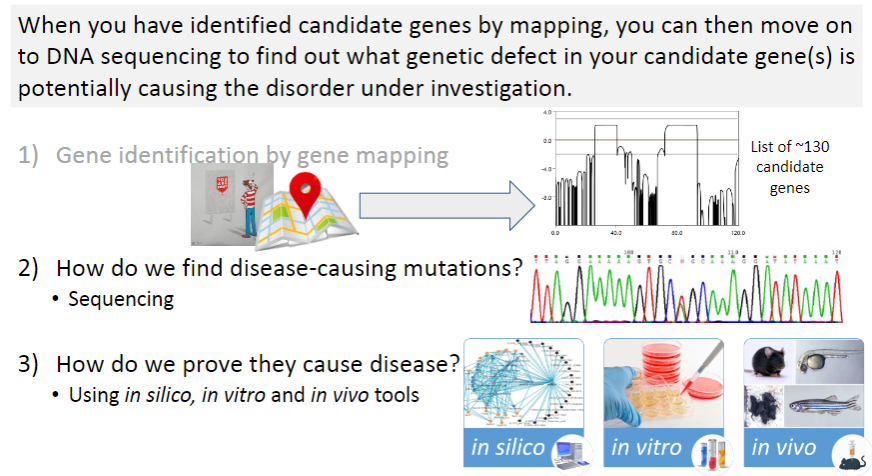
Sequencing: This technique is used to analyze the DNA of candidate genes to find mutations that could potentially cause the disease.
-
After identifying potential mutations, how do researchers confirm that they are actually causing the disease? (3)
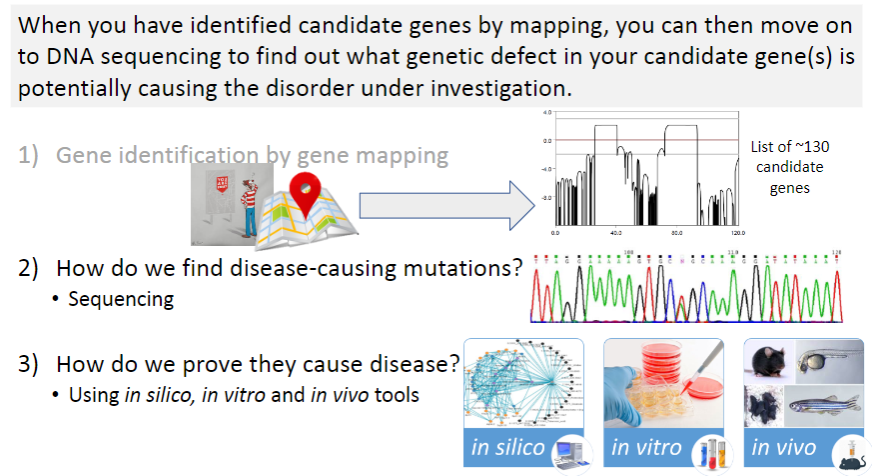
In silico tools: These involve computer-based predictions, such as computational modeling, to assess the potential impact of mutations.
In vitro tools: These are laboratory-based techniques, such as cell culture studies, to observe the effects of mutations in a controlled environment.
In vivo tools: These refer to animal models or clinical studies where mutations are studied in living organisms to observe their effects on disease development.
-
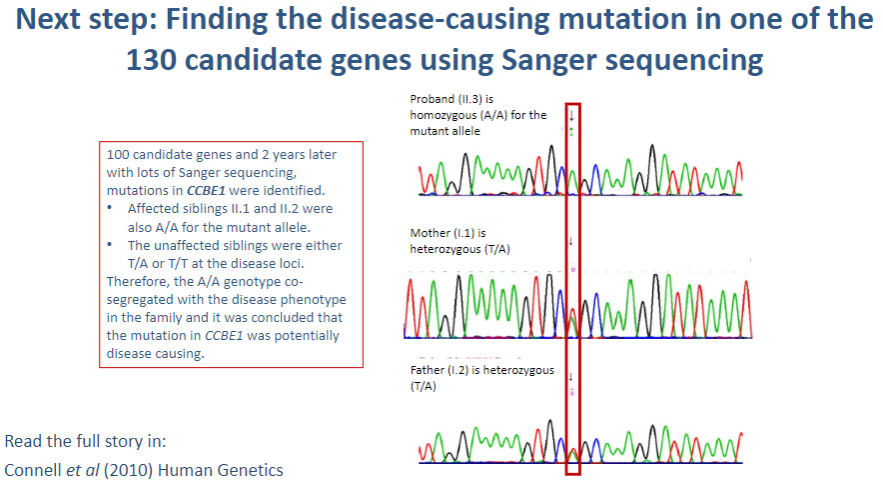
What is the next step after identifying 130 candidate genes in a genetic study? (1)
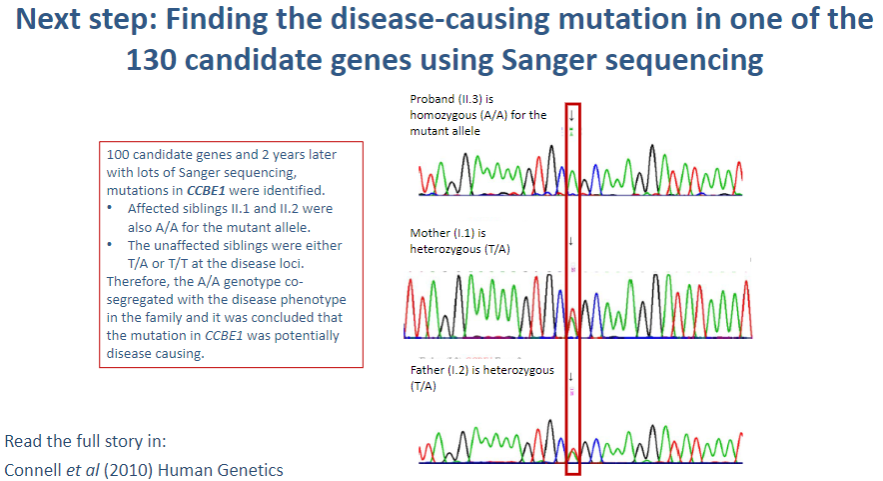
Sanger sequencing: This method is used to sequence the DNA of the candidate genes to identify specific mutations that may be causing the disease.
-
What is Sanger sequencing, and how is it used in identifying disease-causing mutations? (3)
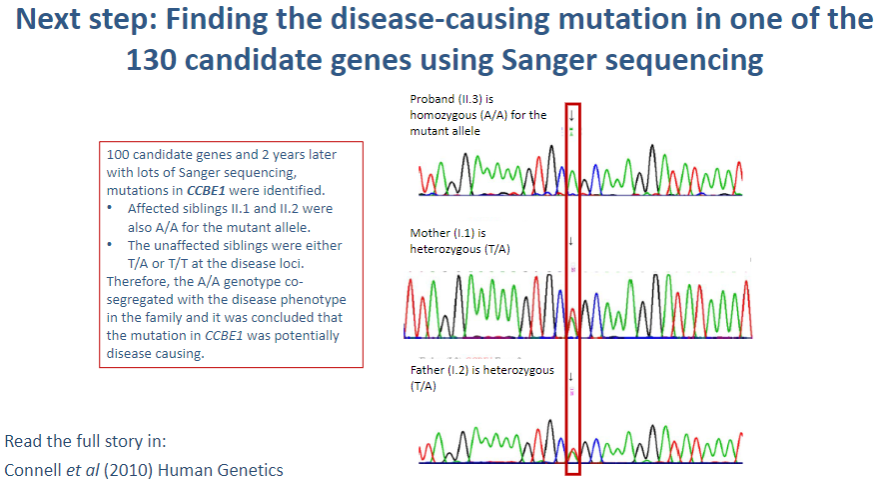
Sanger sequencing: A method of DNA sequencing that uses selective incorporation of chain-terminating dideoxynucleotides.
Process: The gene of interest is amplified by PCR, and the sequence is read using fluorescent markers that indicate which base (A, T, C, G) is incorporated.
Identification of mutations: Any variations in the normal DNA sequence (mutations) can be detected by comparing the obtained sequence to a reference genome.
-
Why is Sanger sequencing a preferred method for confirming disease-causing mutations? (2)
High accuracy: Sanger sequencing provides high-quality, accurate sequences for small regions of DNA, allowing precise identification of mutations.
Targeted sequencing: It allows for focused analysis of specific genes or regions within the genome, such as the 130 candidate genes, making it ideal for finding rare or known mutations associated with diseases.
-
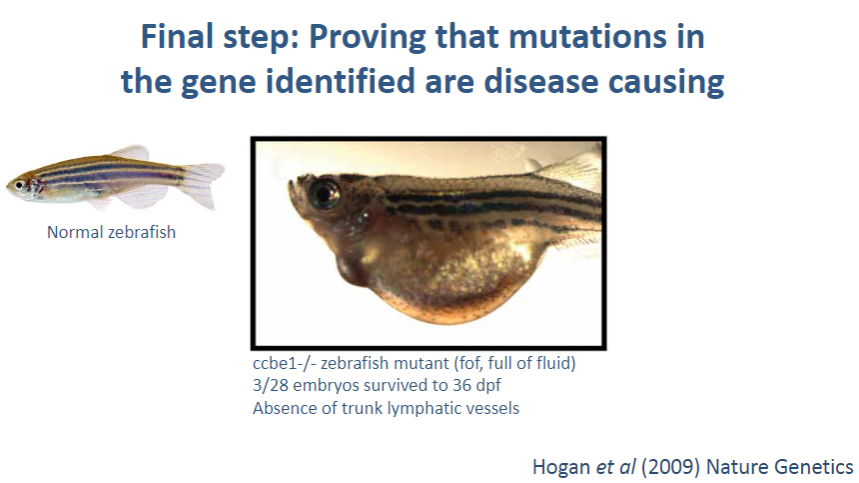
What is the final step in confirming that a mutation in a gene is disease-causing? (1)
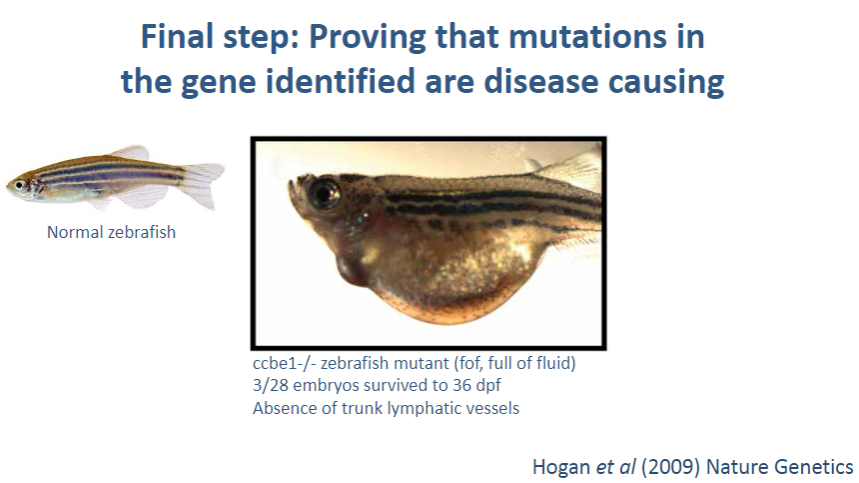
Using in silico, in vitro, and in vivo tools: These methods are used to validate that a mutation in a gene is actually responsible for the disease.
-
What are in silico tools, and how do they help prove that a mutation is disease-causing? (2)
In silico tools: These are computational methods used to predict the effect of genetic mutations on protein function and structure.
Role in proving causality: In silico tools can analyze the mutation's potential to alter the gene’s function or lead to disease by comparing it to known mutation databases and using predictive models.
-
What are in vitro tools, and how do they help confirm a mutation's disease-causing role? (2)
In vitro tools: Laboratory-based experiments (such as cell cultures or protein assays) used to test the effects of the mutation on gene expression or protein function.
Validation: For example, scientists might introduce the mutation into a cell culture model to observe if the mutation disrupts the normal cellular function, suggesting a causal relationship to disease.
-
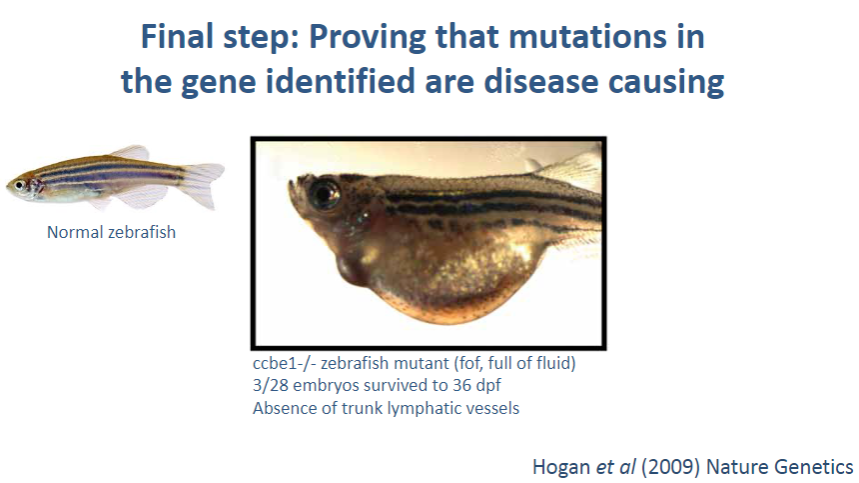
What are in vivo tools, and how do they contribute to proving a mutation causes disease? (2)
In vivo tools: Experiments conducted in living organisms, such as animal models (e.g., mice), that carry the mutation of interest.
Validation: By observing how the mutation affects the organism's phenotype or health, researchers can determine if the mutation leads to the disease, confirming its causality.
-
What is the inheritance pattern of primary lymphoedema? (1)
Autosomal dominant: This form is inherited in an autosomal dominant pattern, meaning one copy of the mutated gene is sufficient to cause the disease.
-
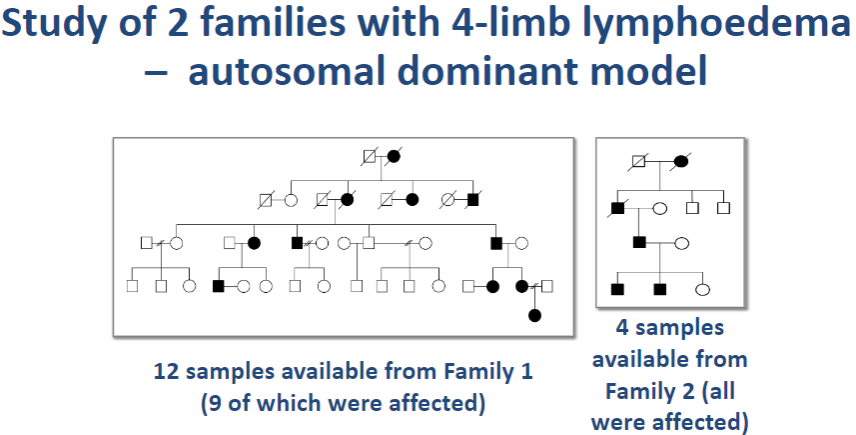
What are the key characteristics of the autosomal dominant form of primary lymphoedema? (4)
4-limb lymphoedema: Affects all four limbs, leading to swelling.
Pubertal/adult onset: The symptoms typically appear during puberty or in adulthood.
Associated with venous incompetence: There is often a co-occurrence of venous problems, such as venous insufficiency.
No other abnormalities: The condition primarily affects the limbs without other major abnormalities.
-
How does traditional linkage analysis help in identifying the cause of primary lymphoedema? (2)
Linkage analysis: This technique helps identify regions of the genome linked to the disease by studying genetic inheritance patterns in families. In primary lymphoedema, it helps to narrow down the possible genomic regions associated with the condition.
-
How does next-generation sequencing contribute to identifying the cause of the disease? (2)
Next-generation sequencing: This advanced method allows for high-throughput, detailed analysis of the genome, helping to pinpoint mutations in genes that might be responsible for the disease.
Gene identification: By sequencing the candidate genes, researchers can directly identify the mutation causing the autosomal dominant form of primary lymphoedema.
-
Picture demonstrating Study of 2 families with 4-limb lymphoedema – autosomal dominant model:
-
How do you generate genotyping data for a linkage analysis study? (2)

Collect DNA samples: Obtain DNA samples from family members, especially affected individuals, and unaffected controls.
Genotyping: Use a genotyping tool, like a SNP array or microarray, to analyze the DNA and generate data on genetic variants (SNPs) across the genome. The data will include information about the alleles present at specific loci for each family member.
-
What is the next step after generating genotyping data? (1)

Create a file with pedigree and genotyping data: Combine the pedigree information (family tree showing relationships and affected status) with the genotyping data to create a file for analysis.
-
How do you run linkage analysis using an autosomal dominant model? (2)
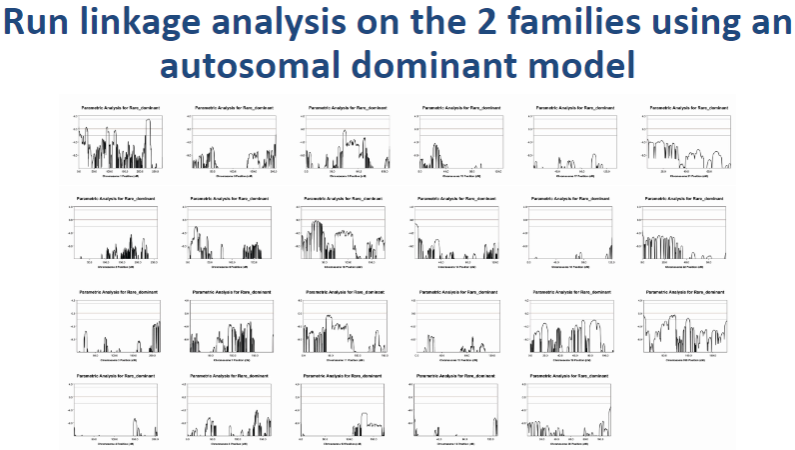
Choose a linkage program: Use software such as Merlin or PLINK to run the linkage analysis.
Set inheritance model: In the software, set the inheritance pattern to autosomal dominant. This will assume that one copy of the mutant allele is sufficient to cause the disease. The program will then compare the genotypes between affected and unaffected family members to identify linked genomic regions.
-
What will linkage analysis identify when using the autosomal dominant model? (1)

Identifying linked regions: The analysis will identify regions of the genome that show consistent inheritance of certain alleles (haplotypes) in affected family members. If a region is linked to the disease, affected individuals will more frequently inherit the same alleles at specific loci.
-
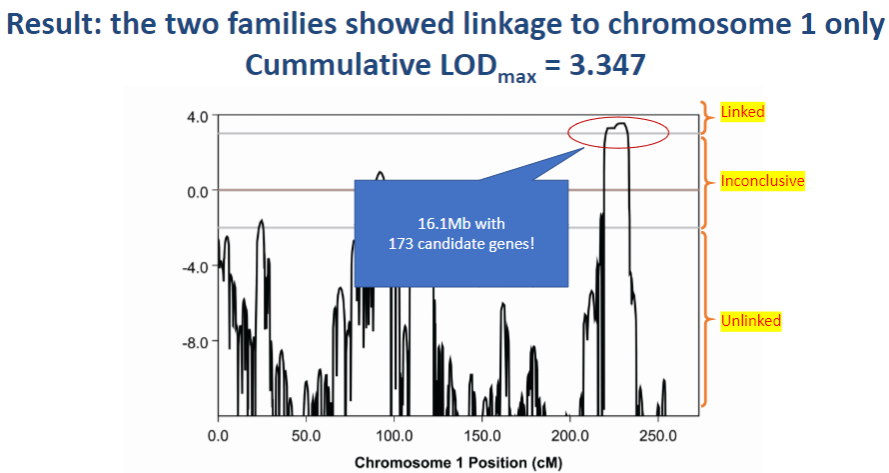
Picture demonstrating Result: the two families showed linkage to chromosome 1 only, Cummulative LODmax = 3.347:
-
How do we find disease-causing mutations? (5)
Traditional Sanger sequencing
Sequences individual genes to identify mutations.
Candidate gene screen
Screens known genes associated with the disease.
Next generation sequencing (NGS)
High-throughput technique for sequencing multiple genes or genomes at once.
Whole genome sequencing (WGS)
Sequences the entire genome, including coding and non-coding regions.
Whole exome sequencing (WES)
Focuses on sequencing only the exons (coding regions) of the genome.
-
What is the advantage of using Next Generation Sequencing (NGS) over traditional methods? (2)
High throughput: NGS can sequence multiple genes or even entire genomes at once, speeding up the process.
Comprehensive: It allows for the detection of a wide range of mutations, including small insertions, deletions, and rare variants that may be missed by traditional methods.
-
When would you use Whole Genome Sequencing (WGS) instead of Whole Exome Sequencing (WES)? (2)
Use WGS when...: You need to examine not only the exons but also the regulatory regions, introns, and other non-coding areas of the genome that may be important for disease.
Use WES when...: You want to focus on the protein-coding regions of the genome, which are more likely to harbor mutations directly related to disease.
-
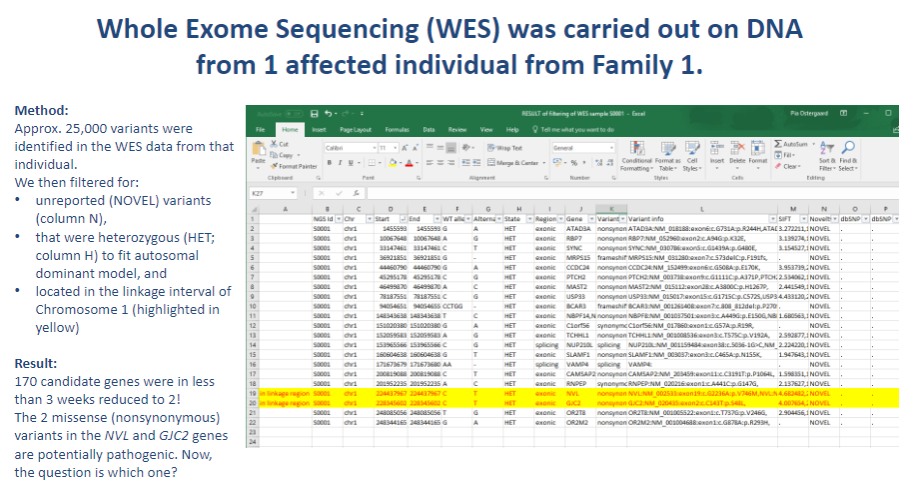
Picture demonstrating Whole Exome Sequencing (WES) was carried out on DNA from 1 affected individual from Family 1:
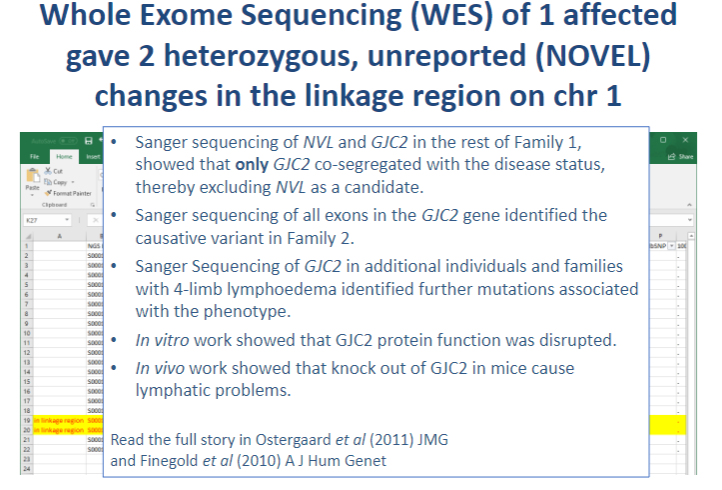
-
What is often the cause of rare diseases? (2)
Mutations in a single gene
Genetic analysis can help identify the causative mutations.
-
What is an example of a technique used to identify causative mutations? (1)
Linkage analysis is an example of such a technique.
-
Why is genetic analysis not always straightforward? (2)
Understanding genetics and the tools applied is necessary.
The process can be complex due to factors like gene interactions, inheritance patterns, and technical limitations.
-
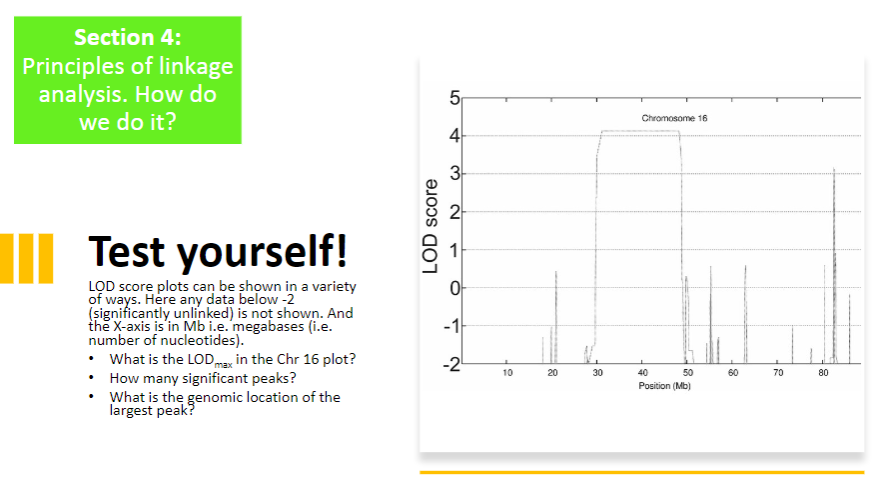
Test Yourself 1:
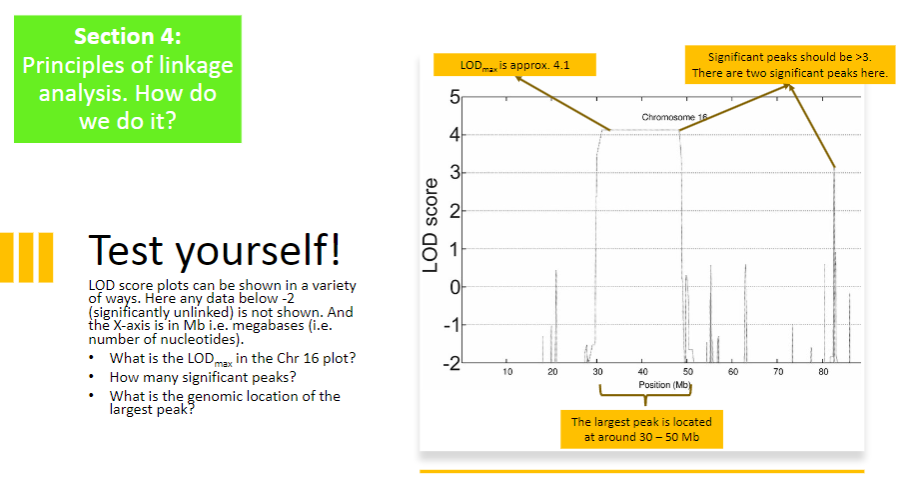
-
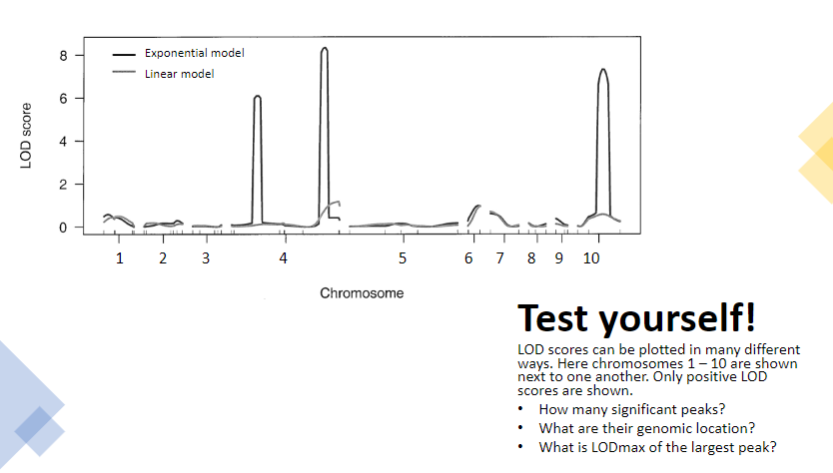
Test yourself 2:
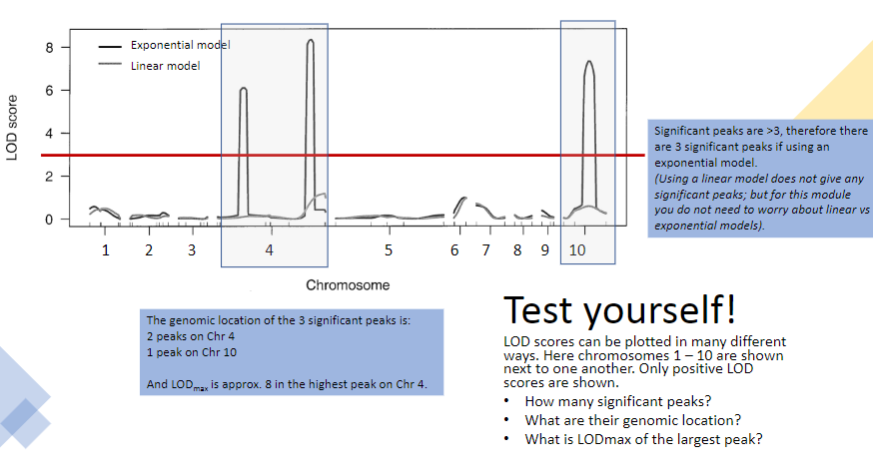
-

Test yourself 3:
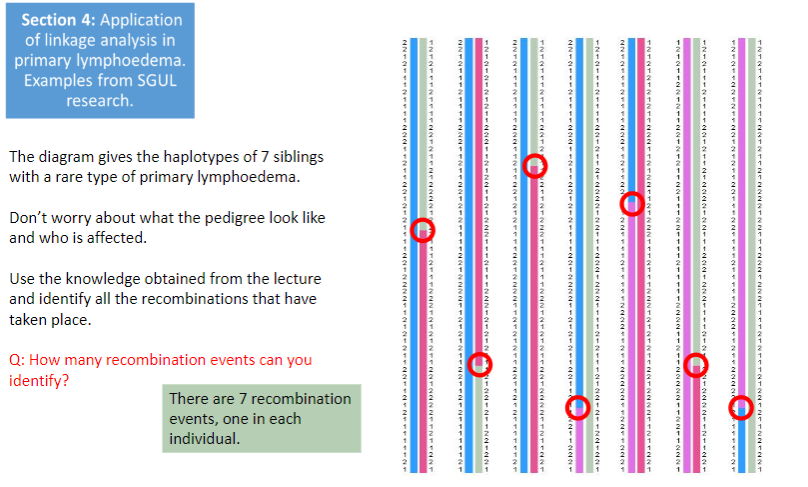
-
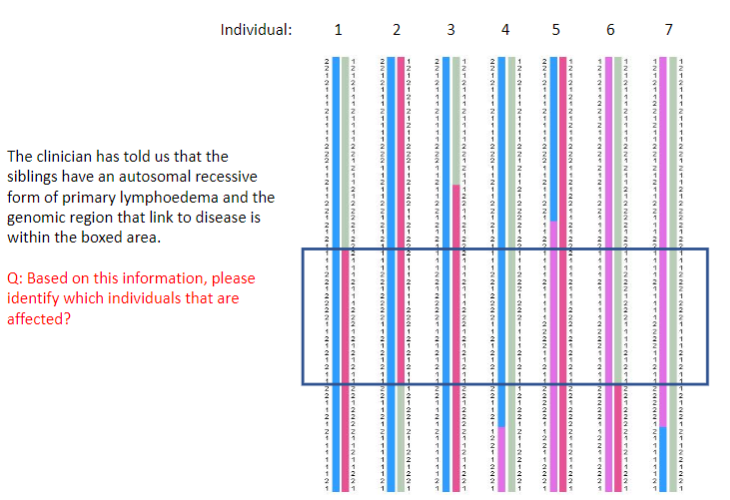
Test yourself 4:
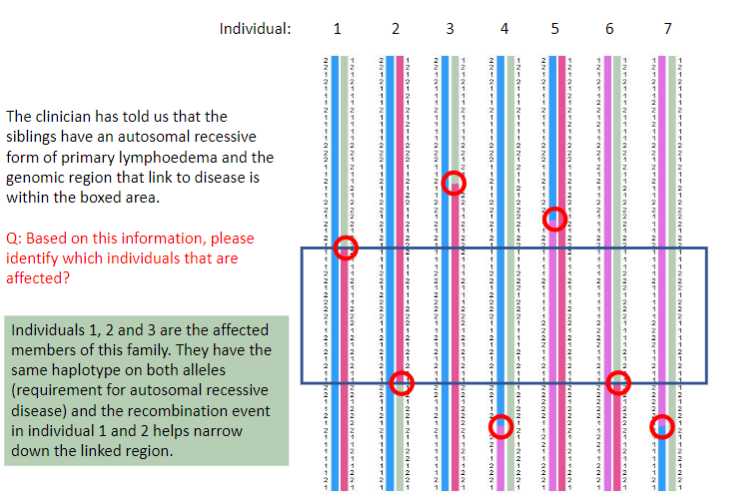
-
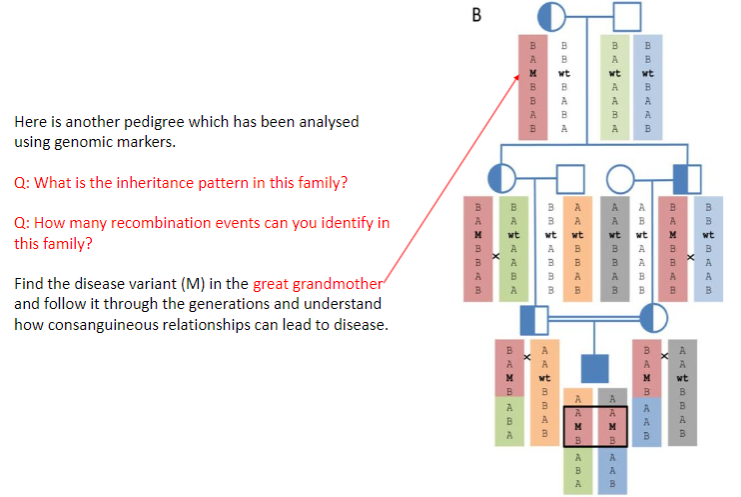
Test yourself 5:
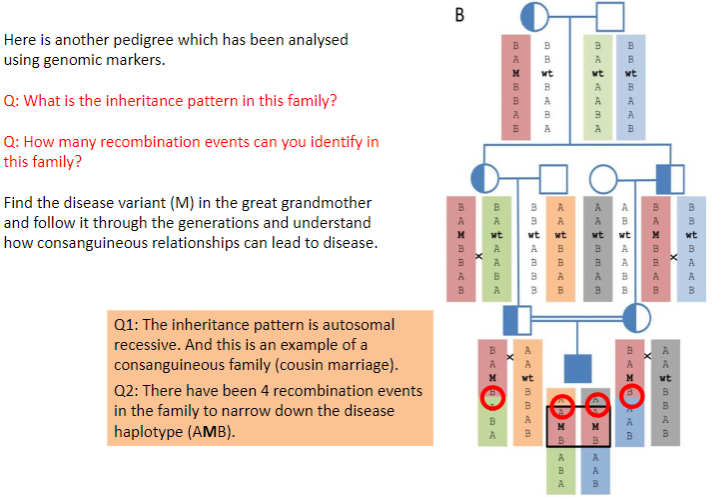
-
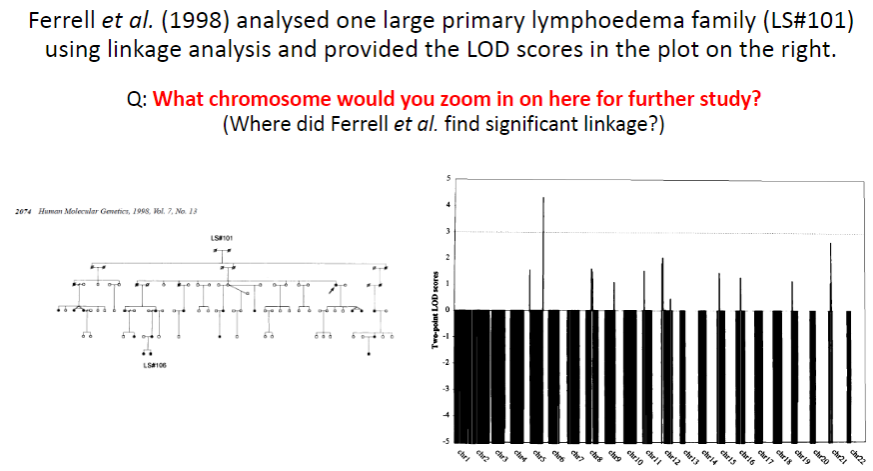
Test yourself 6:
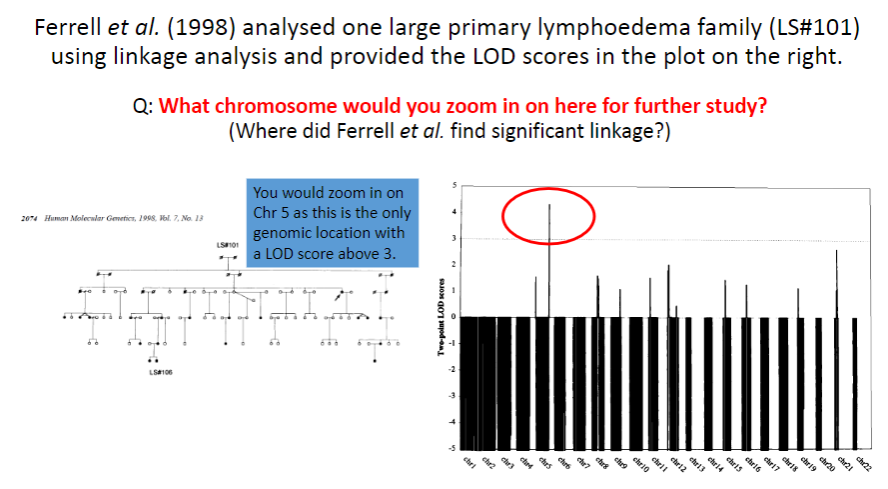
-
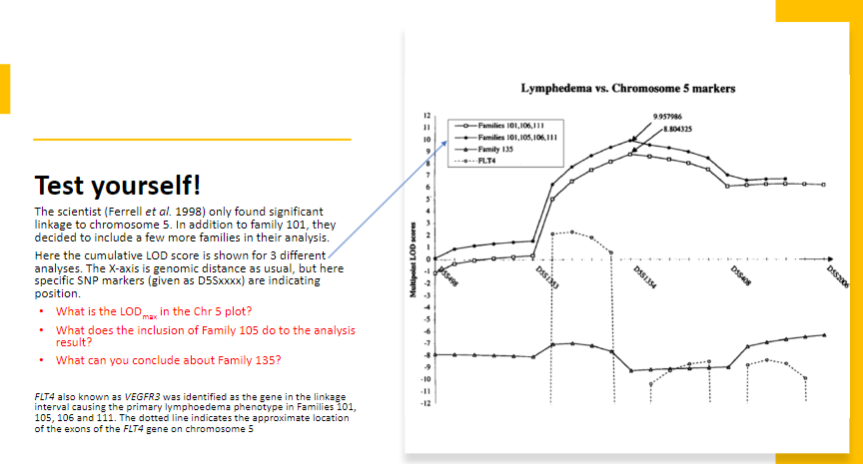
Test yourself 7: