Genome Variation (Genomics)
Genome Variation All the phenotypic variation present in the population is at some level caused by genomic variation. Whether we are talking about a trait such as freckliness, susceptibility to a condition such as alcoholism, risk of developing cancer, blood pressure, or the presence of a monogenic disease such as achondroplasia - all involve genes and differences at the genetic level between individuals. In this session, you will be introduced to some types of common variation: single nucleotide variants; microsatellites; and copy number variation. You should appreciate that it is only actually on rare occasions that these genetic differences we observe lead to severe disease. Learning Outcomes On successful completion of the lecture, students should be able to: Describe how humans differ at the level of their DNA Describe in detail what a single nucleotide variant/polymorphism (SNV/SNP) is, how they are inherited and how common they are Describe how SNVs arise by mismatch repair Describe the difference between a polymorphism and a mutation Describe the evolutionary pressures on a variant Describe in detail what a short tandem repeat (STR or microsatellite) is and how it is inherited Describe how STRs arise due to polymerase slippage, where they are seen in the genome and what effect they might have Describe in detail what a copy number variant is and how it can arise by non-allelic homologous recombination (NAHR)
-
What is the gross structure of the human genome? (1)
23 pairs of chromosomes
-
What is the molecular structure of the human genome? (1)
DNA sequence
-
How many bases are in the human genome? (1)
3 billion bases (3000 Mb)
-
How many genes are estimated to be in the human genome? (1)
~20,000 genes
-
What percentage of the genome codes for protein, known as the exome? (1)
~1.5% of the genome
-
What major macro-level differences are generally associated with disease? (2)
Aneuploidy (occurrence of one or more extra or missing chromosomes in a cell or organism) and translocations (a type of chromosomal abnormality in which a chromosome breaks and a portion of it reattaches to a different chromosome)
-
What micro or molecular-level pathogenic differences can be associated with disease relating to SCA? (2)
-Point mutation and SCA (sickle cell anaemia)
-3 bp deletion in CFTR (cystic fibrosis transmembrane conductance regulator)
-
What type of variants affect traits such as height, hair colour, and intelligence? (1)
Coding variants
-
How much DNA is the same between any two people? (1)
~99.7% DNA is the same
-
How many bases are different between any two people? (1)
~9 million bases different
-
What is considered polymorphic in the genome? (1)
Any position in the genome that varies between individuals is considered polymorphic (a variant)
-
Is every base identical between individuals? (1)

No, two people differ in DNA sequence at ~9 million base pairs (bps).
-
What is a single nucleotide variant (SNV)? (1)
A polymorphism (SNP) where there is a variation in a single nucleotide.
-
What is the frequency of SNVs in the reference genome? (1)
There is 1 SNV every 300 nucleotides.
-
How often does one individual have an SNV? (1)
One individual has 1 SNV every 1000 bases.
-
How many SNVs have been identified in human genomes? (1)
Millions of SNVs have been identified.
-
Where are the majority of SNVs located in the genome? (1)
The majority of SNVs are not in the exome.
-
How are SNVs generated? (1)
SNVs arise when errors during DNA replication are not corrected by the mismatch repair system.
-
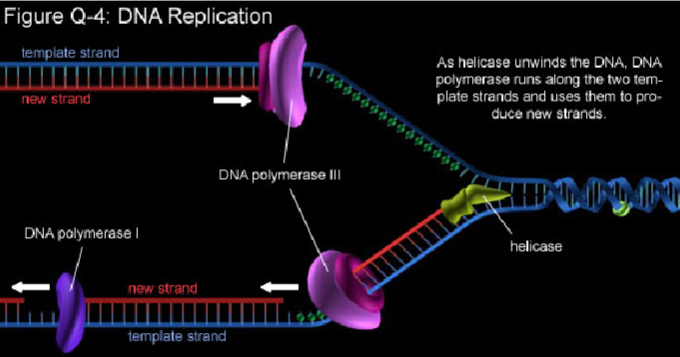
Picture demonstrating the Mismatch repair system:
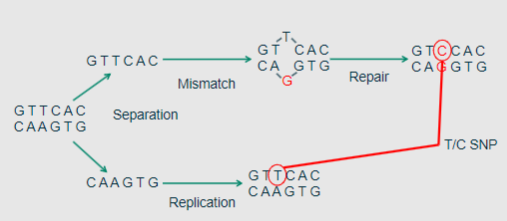
-
Picture demonstrating general nomenclature
-
Where can single nucleotide variants (SNVs) be found? (1)
In genes.
-
What type of SNV results in no amino acid change? (1)
Synonymous SNV.
-
What type of SNV causes an amino acid change? (1)
Non-synonymous (missense) SNV.
-
What type of SNV creates a stop codon? (1)
Nonsense SNV.
-
What role do SNVs play in splice sites? (1)
SNVs can be found in splice sites affecting splicing.
-
How can SNVs affect gene expression? (1)
SNVs can be located in the UTR (untranslated region), influencing gene expression.
-
What role do SNVs play in promoters? (1)
SNVs can be present in promoter regions, impacting gene expression.
-
Can SNVs be found in non-coding regions? (1)
Yes, SNVs can be present in non-coding regions.
-
Why do SNVs not disappear in populations? (1)
Without a deleterious effect or population annihilation, SNVs do not disappear.
-
What is the estimated frequency of the SCA variant allele in Europeans? (1)
0.02%, i.e., 2 in every 10,000 chromosomes.
-
What is the estimated frequency of the SCA variant allele in Africans? (1)
4.5%, i.e., ~1 in every 20 chromosomes.
-
Why is the SCA (Sickle Cell Anaemia) variant allele more frequent in Africa? (2)
It is beneficial in places where malaria is rife (heterozygote advantage).
-
What defines a genetic variant as a polymorphism? (1)
If the minor allele frequency (MAF) > 1% (at least 1 in every 100 chromosomes has the non-reference allele).
-
What are the classifications of polymorphisms based on MAF (Minor Allele Frequency)? (2)
Rare polymorphism: MAF 1-5%.
Common polymorphism: MAF > 5%.
-
What is the safer term to use when discussing genetic differences? (1)
Variant.
-
What is the process through which a new allele arises? (1)
Mutation.
-
How does gene flow contribute to genetic variation? (1)
Migration introduces that variant into another population.
-
What is genetic drift? (1)
Genetic drift is the random fluctuation in the frequency of alleles (both common and rare) in a population from one generation to the next.
-
How does selection affect genetic variants? (1)
It causes non-random change in variant allele frequency due to pathogenic (negative selection) or beneficial (positive selection) effects.
-
When are genetic variants most likely to be neutral? (1)
Depends on their location (in a gene or not), the type of gene, and the specific variant.
-
How many SNVs are estimated to be in the human genome? (1)
Millions.
-
What does an SNV represent? (1)
A position in the genome at which the base can vary.
-
Where can SNVs occur? (1)
Anywhere in the genome (genic or non-genic).
-
What might SNVs affect? (1)
They may do nothing, affect a trait, or be associated with a disorder.
-
How are SNVs generally categorized? (1)
Generally bi-allelic.
-
What causes SNVs? (1)
They are caused by mutation and mismatch repair.
-
What are pathogenic SNVs also called? (1)
Pathogenic SNVs are often referred to as disease-causing mutations or disease-associated variants.
-
Is every base identical between individuals? (1)

No, two people differ in DNA sequence at ~9 million bases.
-
Is every genome exactly 3000Mb? (1)

No, while the human genome is approximately 3000Mb, there can be variations among individuals.
-
What is a microsatellite? (1)

A microsatellite is also known as a short tandem repeat.
-

What does the "AC" in a microsatellite represent? (1)

The "AC" represents the repeat unit, which is repeated in tandem (one after another).
-
Picture demonstrating microsatellites further:
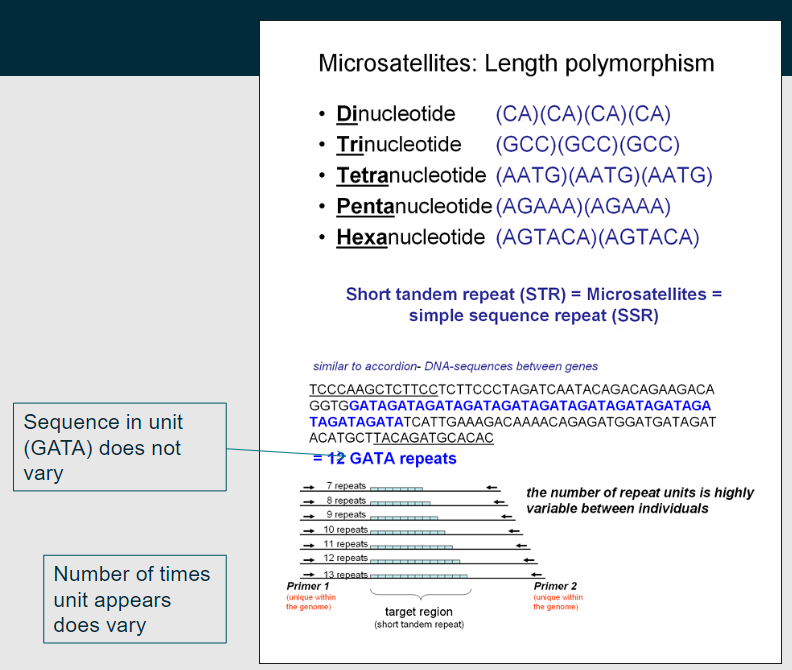
-
Picture demonstrating the different genotypes in a family through microsatellites:
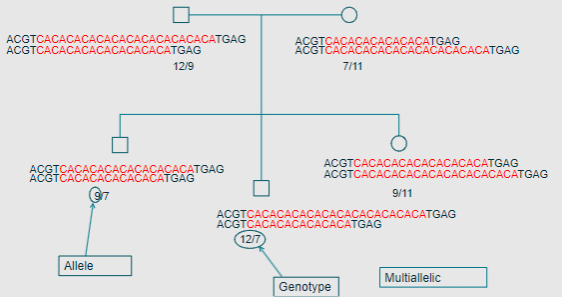
-
What is the polymerase slippage model? (1)
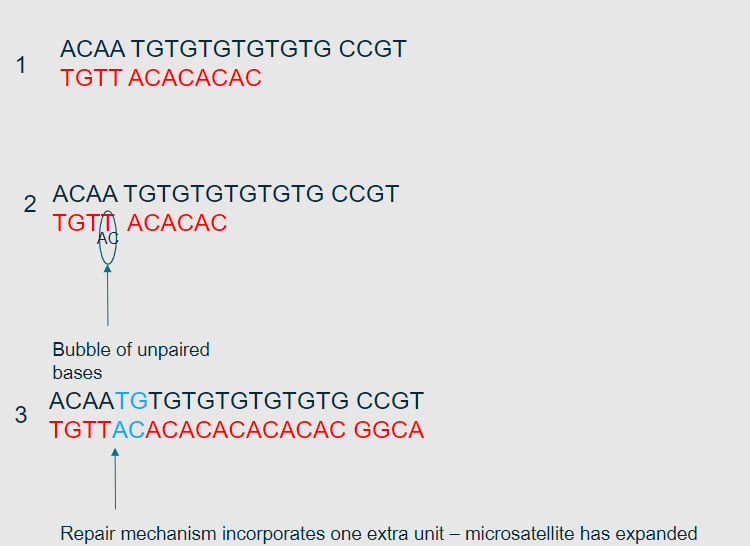
It is a model explaining how errors occur during DNA replication due to the slippage of DNA polymerase on repetitive sequences.
-
How does polymerase slippage lead to replication errors? (2)
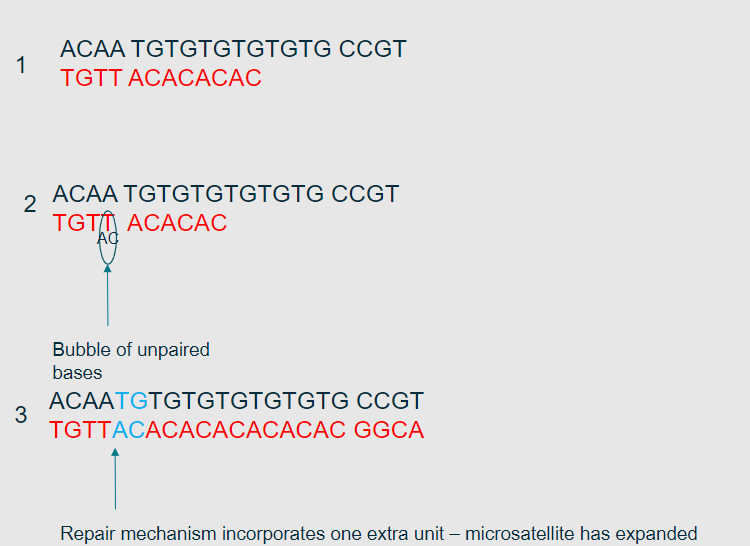
Polymerase may insert extra bases or skip bases while replicating repetitive sequences.
This results in insertions or deletions (indels) in the DNA sequence.
-
What types of sequences are most affected by polymerase slippage? (1)
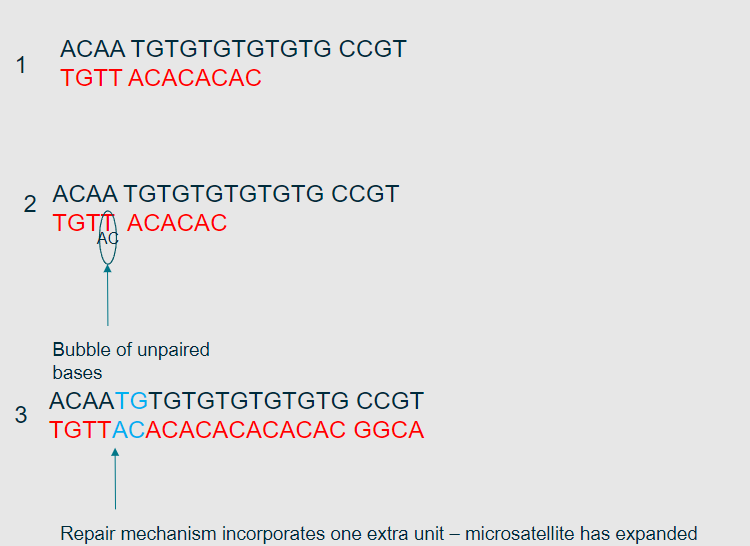
Repetitive sequences, such as microsatellites, are most affected by polymerase slippage.
-
Picture further demonstrating the Polymerase slippage model?
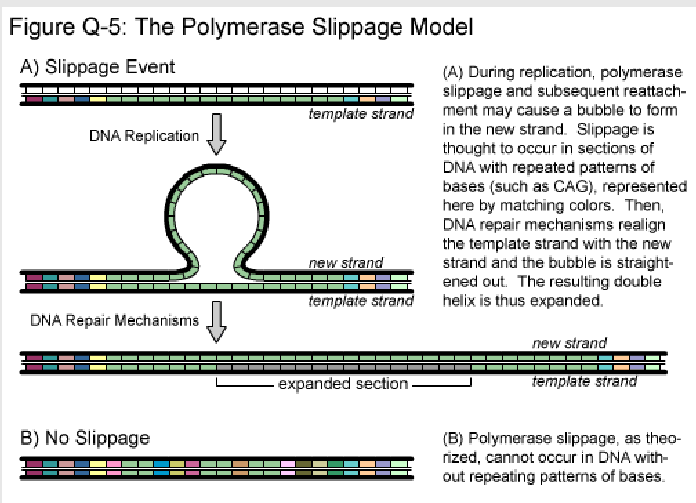
-
Where can microsatellites be found in the genome? (4)
In regions not coding for protein.
In intronic or UTR regions, which may affect gene expression.
In intergenic regions.
In exonic regions, potentially leading to extra amino acids in proteins.
-
What is a pathogenic example of a microsatellite disorder? (1)
Expansion disorders, such as Huntington’s disease, which is a trinucleotide repeat expansion disorder.
-
What are key characteristics of microsatellites? (3)
Tandem Repeats: Microsatellites consist of short tandem repeat sequences (1-6 base pairs), such as CACACA or AGAGAG, repeated multiple times.
Highly Polymorphic: The number of repeats can vary greatly among individuals, making them multiallelic and useful in genetic profiling.
Genomic Distribution: They are distributed throughout the genome, located in coding regions, non-coding regions, and regulatory elements, and can affect function depending on their location.
-
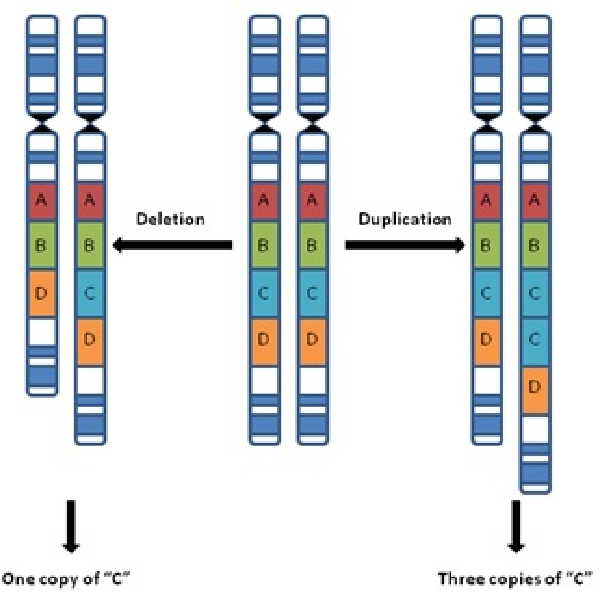
Picture example of a Copy Number Variant (CNV)
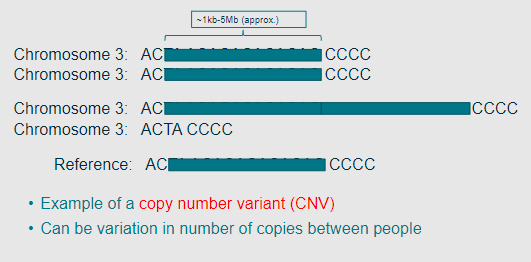
-
Another picture demonstration Copy Number Variation:

-
What is copy number variation? (1)
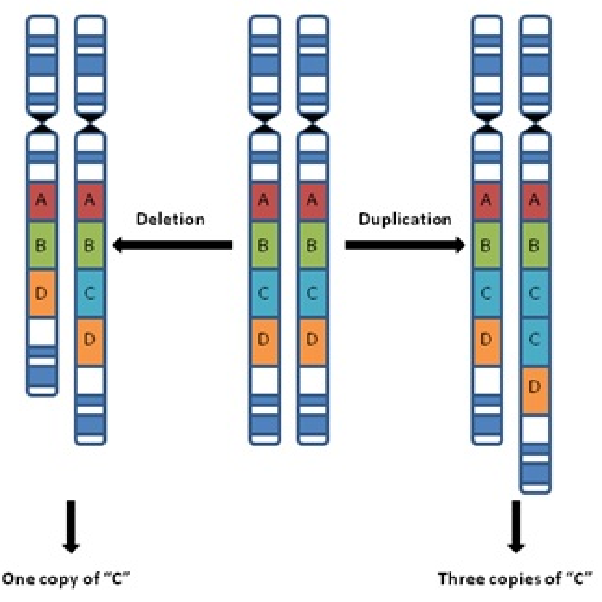
Variation in the number of copies of a particular gene or genomic region (1)
-
What does a pair of homologous chromosomes consist of? (1)
Two copies of each chromosome, e.g., two copies of chromosome 12 (1)
-
In theory, how many copies of every locus (gene, base, genomic region) are present? (1)
Every locus is present as diploid
-
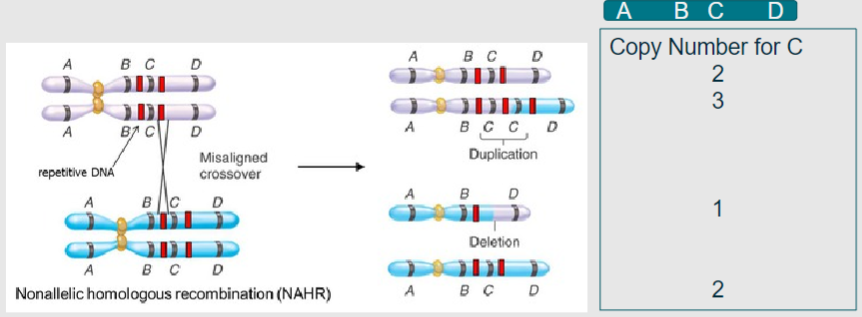
What is non-allelic homologous recombination? (2)
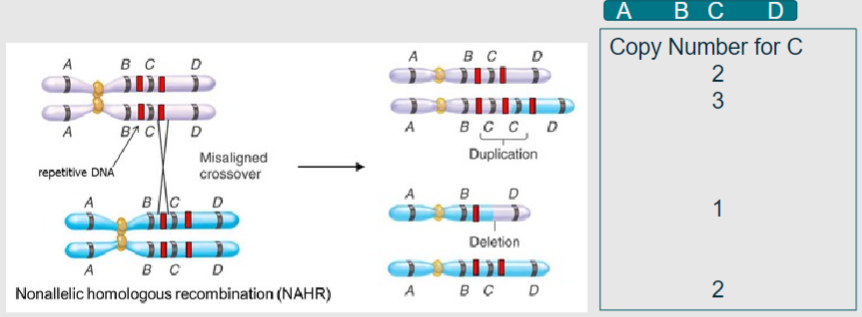
A process in meiosis where homologous chromosomes align, leading to duplication or deletion of genetic material
Results in copy number variation instead of the beneficial shuffling of alleles
-
What do the grey and blue represent in the context of homologous chromosomes during meiosis? (1)
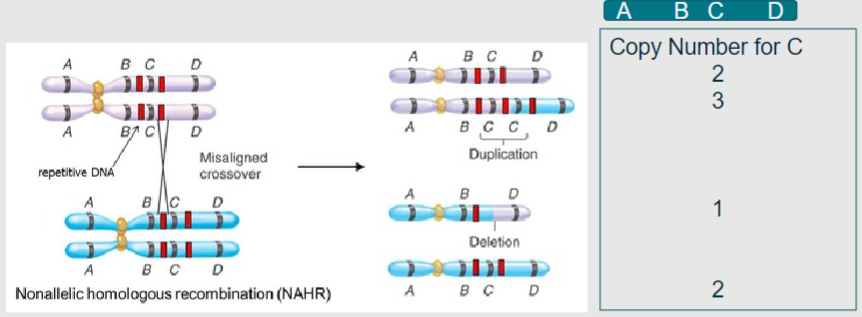
Grey and blue represent homologous chromosomes aligning in meiosis I (1)
-
What do the red bands indicate during non-allelic homologous recombination? (1)
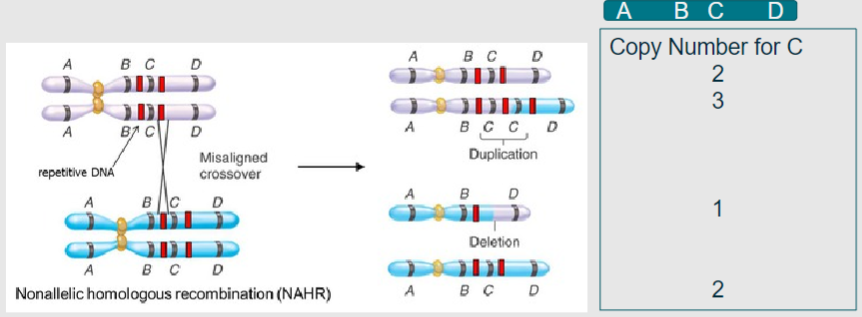
Regions of high sequence similarity, often derived from viral or bacterial genomes that have been incorporated through evolution (1)
-
What are CNVs (Copy Number Variants)? (2)
CNVs can be intergenic or affect one or more genes.
They are typically quite large, greater than 1kb
-
What percentage of the genome is estimated to be CNV? (1)
Approximately 12% of the genome.
-
How many CNVs have been identified? (1)
Over 2000 CNVs have been identified.
-
What is the size range for CNVs? (1)
CNVs range from 1kb to 5000kb.
-
Can you give an example of a pathogenic copy number variation? (1)
Microdeletion disorders, such as DiGeorge syndrome.
-
What are the types of common genetic variants? (2)
Single Nucleotide Polymorphisms (SNPs) - approximately 17 million identified.
Microsatellites - about 3% of the genome.
-
What is the approximate number of CNVs identified per genome? (1)
Around 100 CNVs per genome
-
What does it mean that everyone “has” every variant? (1)
It means that all individuals carry some genetic variants, but the difference between individuals lies in the specific combination of alleles (genotype) they have at each variant.
-
What is common regarding genetic variants in the genome? (2)
Many variants are present throughout the genome.
If biallelic, the frequency of the minor allele is relatively high.
-
What is the population frequency? (1)
It is the proportion of chromosomes that carry each allele in the population.
-
What is the association between common variants and diseases or traits? (3)
Most common variants do not cause Mendelian, monogenic disorders.
The majority are probably neutral, particularly intergenic variants.
However, they may impact complex, non-Mendelian disorders and contribute to individual variation.
-
What are the potential effects of genetic variants? (4)
They can be beneficial.
They can be pathogenic.
Most are neutral.
Variants can be used as markers to help find disease-causing genes and mutations.
-
What methodologies can be used for genetic analysis involving variants? (2)
Autozygosity mapping and linkage studies using microsatellites and SNPs.
Association analysis using SNPs and CNVs.
-
Picture demonstrating the book analogy:
-
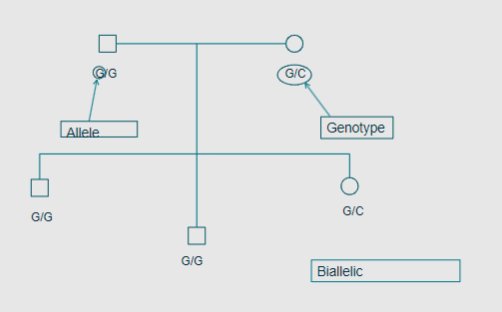
What is a locus in the context of the genome? (2)
A locus is a unique position in the genome.
It can refer to a single base or an entire genomic region.
-
What is an allele? (2)
An allele is a particular form of a specific locus.
It can also range from a single base to an entire genomic region.
-
What is the genotype of an individual regarding autosomal loci? (2)
An individual has 2 alleles for any autosomal locus.
The genotype can be heterozygous or homozygous
-
What does biallelic mean? (1)
Biallelic means there are 2 possible alleles at a locus.
-
What does triallelic mean? (1)
Triallelic means there are 3 possible alleles at a locus.
-
What does multiallelic mean? (1)
Multiallelic means there are more than 3 possible alleles at a locus.
-
How is the presence of an allele expressed in a population? (1)
The presence of an allele is expressed as a frequency or percentage.
-
Do two populations of the same species need to have the same frequency at the same locus? (1)
No, two populations of the same species need not have the same frequency at the same locus.

